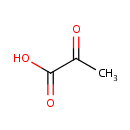
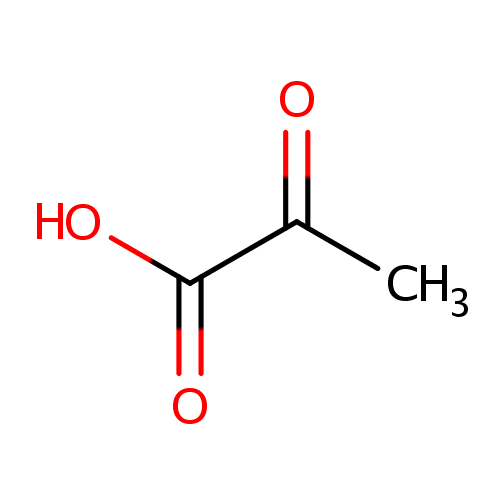
Pyruvic acid (PAMDB000102)
Enzymes
- General function:
- Involved in magnesium ion binding
- Specific function:
- 2 pyruvate = 2-acetolactate + CO(2)
- Gene Name:
- ilvI
- Locus Tag:
- PA4696
- Molecular weight:
- 63 kDa
Reactions
| 2 pyruvate = 2-acetolactate + CO(2). |
- General function:
- Involved in acetolactate synthase activity
- Specific function:
- 2 pyruvate = 2-acetolactate + CO(2)
- Gene Name:
- ilvH
- Locus Tag:
- PA4695
- Molecular weight:
- 17.8 kDa
Reactions
| 2 pyruvate = 2-acetolactate + CO(2). |
- General function:
- Involved in biosynthetic process
- Specific function:
- Chorismate + L-glutamine = anthranilate + pyruvate + L-glutamate
- Gene Name:
- trpE
- Locus Tag:
- PA0609
- Molecular weight:
- 54.6 kDa
Reactions
| Chorismate + L-glutamine = anthranilate + pyruvate + L-glutamate. |
- General function:
- Involved in anthranilate phosphoribosyltransferase activity
- Specific function:
- Chorismate + L-glutamine = anthranilate + pyruvate + L-glutamate
- Gene Name:
- trpD
- Locus Tag:
- PA0650
- Molecular weight:
- 37.4 kDa
Reactions
| Chorismate + L-glutamine = anthranilate + pyruvate + L-glutamate. |
| N-(5-phospho-D-ribosyl)-anthranilate + diphosphate = anthranilate + 5-phospho-alpha-D-ribose 1-diphosphate. |
- General function:
- Involved in D-serine ammonia-lyase activity
- Specific function:
- D-serine = pyruvate + NH(3)
- Gene Name:
- dsdA
- Locus Tag:
- PA3357
- Molecular weight:
- 48.2 kDa
Reactions
| D-serine = pyruvate + NH(3). |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the formation of alpha-ketobutyrate from threonine in a two-step reaction. The first step is a dehydration of threonine, followed by rehydration and liberation of ammonia. Deaminates L-threonine, but also L-serine to a lesser extent
- Gene Name:
- ilvA
- Locus Tag:
- PA1326
- Molecular weight:
- 55.9 kDa
Reactions
| L-threonine = 2-oxobutanoate + NH(3). |
- General function:
- Involved in transferase activity, transferring acyl groups
- Specific function:
- The pyruvate dehydrogenase complex catalyzes the overall conversion of pyruvate to acetyl-CoA and CO(2). It contains multiple copies of three enzymatic components:pyruvate dehydrogenase (E1), dihydrolipoamide acetyltransferase (E2) and lipoamide dehydrogenase (E3)
- Gene Name:
- aceF
- Locus Tag:
- PA5016
- Molecular weight:
- 56.7 kDa
Reactions
| Acetyl-CoA + enzyme N(6)-(dihydrolipoyl)lysine = CoA + enzyme N(6)-(S-acetyldihydrolipoyl)lysine. |
- General function:
- Involved in magnesium ion binding
- Specific function:
- Pyruvate + ferricytochrome b1 + H(2)O = acetate + CO(2) + ferrocytochrome b1
- Gene Name:
- poxB
- Locus Tag:
- PA5297
- Molecular weight:
- 62.3 kDa
Reactions
| Pyruvate + ubiquinone + H(2)O = acetate + CO(2) + ubiquinol. |
- General function:
- Involved in protein-N(PI)-phosphohistidine-sugar phosphotransferase activity
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (sugar PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitantly with their translocation across the cell membrane. This system is involved in N-acetylglucosamine transport
- Gene Name:
- nagE
- Locus Tag:
- PA3761
- Molecular weight:
- 60.6 kDa
Reactions
| Protein EIIA N(pi)-phospho-L-histidine + protein EIIB = protein EIIA + protein EIIB N(pi)-phospho-L-histidine/cysteine. |
| Protein EIIB N(pi)-phospho-L-histidine/cysteine + sugar = protein EIIB + sugar phosphate. |
- General function:
- Involved in D-amino-acid dehydrogenase activity
- Specific function:
- Oxidative deamination of D-amino acids
- Gene Name:
- dadA
- Locus Tag:
- PA5304
- Molecular weight:
- 47.1 kDa
Reactions
| A D-amino acid + H(2)O + acceptor = a 2-oxo acid + NH(3) + reduced acceptor. |
- General function:
- Involved in catalytic activity
- Specific function:
- L-aspartate 4-semialdehyde + pyruvate = dihydrodipicolinate + 2 H(2)O
- Gene Name:
- dapA
- Locus Tag:
- PA1010
- Molecular weight:
- 31.4 kDa
Reactions
| L-aspartate 4-semialdehyde + pyruvate = dihydrodipicolinate + 2 H(2)O. |
- General function:
- Involved in catalytic activity
- Specific function:
- Interconversion of serine and glycine
- Gene Name:
- glyA
- Locus Tag:
- PA4602
- Molecular weight:
- 45.2 kDa
Reactions
| 5,10-methylenetetrahydrofolate + glycine + H(2)O = tetrahydrofolate + L-serine. |
- General function:
- Involved in magnesium ion binding
- Specific function:
- ATP + pyruvate = ADP + phosphoenolpyruvate
- Gene Name:
- pykF
- Locus Tag:
- PA1498
- Molecular weight:
- 51.5 kDa
Reactions
| ATP + pyruvate = ADP + phosphoenolpyruvate. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- The pyruvate dehydrogenase complex catalyzes the overall conversion of pyruvate to acetyl-CoA and CO(2). It contains multiple copies of three enzymatic components:pyruvate dehydrogenase (E1), dihydrolipoamide acetyltransferase (E2) and lipoamide dehydrogenase (E3)
- Gene Name:
- aceE
- Locus Tag:
- PA5015
- Molecular weight:
- 99.6 kDa
Reactions
| Pyruvate + [dihydrolipoyllysine-residue acetyltransferase] lipoyllysine = [dihydrolipoyllysine-residue acetyltransferase] S-acetyldihydrolipoyllysine + CO(2). |
- General function:
- Involved in L-serine ammonia-lyase activity
- Specific function:
- Deaminates also threonine, particularly when it is present in high concentration
- Gene Name:
- sdaA
- Locus Tag:
- PA2443
- Molecular weight:
- 48.9 kDa
Reactions
| L-serine = pyruvate + NH(3). |
- General function:
- Involved in sugar:hydrogen symporter activity
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (sugar PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitantly with their translocation across the cell membrane. This system is involved in fructose transport
- Gene Name:
- fruA
- Locus Tag:
- PA3560
- Molecular weight:
- 59 kDa
Reactions
| Protein EIIB N(pi)-phospho-L-histidine/cysteine + sugar = protein EIIB + sugar phosphate. |
- General function:
- Involved in magnesium ion binding
- Specific function:
- ATP + pyruvate = ADP + phosphoenolpyruvate
- Gene Name:
- pykA
- Locus Tag:
- PA4329
- Molecular weight:
- 52.3 kDa
Reactions
| ATP + pyruvate = ADP + phosphoenolpyruvate. |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the phosphorylation of pyruvate to phosphoenolpyruvate
- Gene Name:
- ppsA
- Locus Tag:
- PA1770
- Molecular weight:
- 85.8 kDa
Reactions
| ATP + pyruvate + H(2)O = AMP + phosphoenolpyruvate + phosphate. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- (S)-malate + NAD(+) = pyruvate + CO(2) + NADH
- Gene Name:
- sfcA
- Locus Tag:
- PA3471
- Molecular weight:
- 62.4 kDa
Reactions
| (S)-malate + NAD(+) = pyruvate + CO(2) + NADH. |
- General function:
- Involved in 4-amino-4-deoxychorismate lyase activity
- Specific function:
- Converts 4-amino-4-deoxychorismate into 4-aminobenzoate (PABA) and pyruvate
- Gene Name:
- pabC
- Locus Tag:
- PA2964
- Molecular weight:
- 29.9 kDa
Reactions
| 4-amino-4-deoxychorismate = 4-aminobenzoate + pyruvate. |
- General function:
- Involved in L-serine ammonia-lyase activity
- Specific function:
- Deaminates also threonine, particularly when it is present in high concentration
- Gene Name:
- sdaB
- Locus Tag:
- PA5379
- Molecular weight:
- 49.2 kDa
Reactions
| L-serine = pyruvate + NH(3). |
- General function:
- Involved in catalytic activity
- Specific function:
- (S)-lactate + 2 ferricytochrome c = pyruvate + 2 ferrocytochrome c + 2 H(+)
- Gene Name:
- lldD
- Locus Tag:
- PA4771
- Molecular weight:
- 41.1 kDa
Reactions
| (S)-lactate + 2 ferricytochrome c = pyruvate + 2 ferrocytochrome c + 2 H(+). |
- General function:
- Involved in protein binding
- Specific function:
- Component of the phosphoenolpyruvate-dependent nitrogen- metabolic phosphotransferase system (nitrogen-metabolic PTS), that seems to be involved in regulating nitrogen metabolism. Enzyme I- Ntr transfers the phosphoryl group from phosphoenolpyruvate (PEP) to the phosphoryl carrier protein (NPr). Could function in the transcriptional regulation of sigma-54 dependent operons in conjunction with the NPr (ptsO) and EIIA-Ntr (ptsN) proteins
- Gene Name:
- ptsP
- Locus Tag:
- PA0337
- Molecular weight:
- 83.6 kDa
Reactions
| Phosphoenolpyruvate + protein L-histidine = pyruvate + protein N(pi)-phospho-L-histidine. |
- General function:
- Involved in sugar:hydrogen symporter activity
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitant with their translocation across the cell membrane. This system is involved in galactitol transport
- Gene Name:
- gatB
- Locus Tag:
- PA4484
- Molecular weight:
- 53.1 kDa
Reactions
| Protein EIIB N(pi)-phospho-L-histidine/cysteine + sugar = protein EIIB + sugar phosphate. |
- General function:
- Involved in oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor
- Specific function:
- Fermentative lactate dehydrogenase
- Gene Name:
- ldhA
- Locus Tag:
- PA0927
- Molecular weight:
- 35.8 kDa
Reactions
| (R)-lactate + NAD(+) = pyruvate + NADH. |
- General function:
- Involved in transporter activity
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (sugar PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitantly with their translocation across the cell membrane. This system is involved in galactitol transport
- Gene Name:
- gatA
- Locus Tag:
- PA4483
- Molecular weight:
- 51.9 kDa
Reactions
| Protein EIIA N(pi)-phospho-L-histidine + protein EIIB = protein EIIA + protein EIIB N(pi)-phospho-L-histidine/cysteine. |
- General function:
- Involved in lyase activity
- Specific function:
- Catalyzes the cleavage of L-allo-threonine and L- threonine to glycine and acetaldehyde. L-threo-phenylserine and L- erythro-phenylserine are also good substrates
- Gene Name:
- ltaE
- Locus Tag:
- PA0902
- Molecular weight:
- 35.4 kDa
Reactions
| L-threonine = glycine + acetaldehyde. |
| L-allo-threonine = glycine + acetaldehyde. |
- General function:
- Involved in carbon-carbon lyase activity
- Specific function:
- Catalyzes the reversible retro-aldol cleavage of 2-keto- 3-deoxy-L-rhamnonate (KDR) to pyruvate and lactaldehyde. 2-keto-3- deoxy-L-mannonate, 2-keto-3-deoxy-L-lyxonate and 4-hydroxy-2- ketoheptane-1,7-dioate (HKHD) are also reasonably good substrates, although 2-keto-3-deoxy-L-rhamnonate is likely to be the physiological substrate
- Gene Name:
- rhmA
- Locus Tag:
- PA4128
- Molecular weight:
- 28.2 kDa
Reactions
| 2-dehydro-3-deoxy-L-rhamnonate = pyruvate + (R)-lactaldehyde. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- (S)-malate + NADP(+) = pyruvate + CO(2) + NADPH
- Gene Name:
- maeB
- Locus Tag:
- PA5046
- Molecular weight:
- 45.4 kDa
Reactions
| (S)-malate + NADP(+) = pyruvate + CO(2) + NADPH. |
- General function:
- Involved in 1-deoxy-D-xylulose-5-phosphate synthase activity
- Specific function:
- Catalyzes the acyloin condensation reaction between C atoms 2 and 3 of pyruvate and glyceraldehyde 3-phosphate to yield 1-deoxy-D-xylulose-5-phosphate (DXP)
- Gene Name:
- dxs
- Locus Tag:
- PA4044
- Molecular weight:
- 68 kDa
Reactions
| Pyruvate + D-glyceraldehyde 3-phosphate = 1-deoxy-D-xylulose 5-phosphate + CO(2). |
- General function:
- Involved in catalytic activity
- Specific function:
- Catalyzes the formation of pyruvate and succinate from 2-methylisocitrate
- Gene Name:
- prpB
- Locus Tag:
- PA0796
- Molecular weight:
- 32.1 kDa
Reactions
| (2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate = pyruvate + succinate. |
- General function:
- Involved in phosphoenolpyruvate-dependent sugar phosphotransferase system
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitant with their translocation across the cell membrane. This system is involved in galactitol transport
- Gene Name:
- gatC
- Locus Tag:
- PA4482
- Molecular weight:
- 10.5 kDa
- General function:
- Involved in 1-aminocyclopropane-1-carboxylate synthase activity
- Specific function:
- Specific function unknown
- Gene Name:
- yfbQ
- Locus Tag:
- PA2828
- Molecular weight:
- 44.8 kDa
Reactions
| L-alanine + 2-oxoglutarate = pyruvate + L-glutamate. |
- General function:
- Amino acid transport and metabolism
- Specific function:
- Specific function unknown
- Gene Name:
- yfdZ
- Locus Tag:
- PA4715
- Molecular weight:
- 46.1 kDa
Reactions
| L-alanine + 2-oxoglutarate = pyruvate + L-glutamate. |
Transporters
- General function:
- Involved in protein-N(PI)-phosphohistidine-sugar phosphotransferase activity
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (sugar PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitantly with their translocation across the cell membrane. This system is involved in N-acetylglucosamine transport
- Gene Name:
- nagE
- Locus Tag:
- PA3761
- Molecular weight:
- 60.6 kDa
Reactions
| Protein EIIA N(pi)-phospho-L-histidine + protein EIIB = protein EIIA + protein EIIB N(pi)-phospho-L-histidine/cysteine. |
| Protein EIIB N(pi)-phospho-L-histidine/cysteine + sugar = protein EIIB + sugar phosphate. |
- General function:
- Involved in sugar:hydrogen symporter activity
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (sugar PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitantly with their translocation across the cell membrane. This system is involved in fructose transport
- Gene Name:
- fruA
- Locus Tag:
- PA3560
- Molecular weight:
- 59 kDa
Reactions
| Protein EIIB N(pi)-phospho-L-histidine/cysteine + sugar = protein EIIB + sugar phosphate. |
- General function:
- Involved in phosphoenolpyruvate-dependent sugar phosphotransferase system
- Specific function:
- The phosphoenolpyruvate-dependent sugar phosphotransferase system (PTS), a major carbohydrate active -transport system, catalyzes the phosphorylation of incoming sugar substrates concomitant with their translocation across the cell membrane. This system is involved in galactitol transport
- Gene Name:
- gatC
- Locus Tag:
- PA4482
- Molecular weight:
- 10.5 kDa