| Identification |
| Name: |
D-amino acid dehydrogenase small subunit |
| Synonyms: |
Not Available |
| Gene Name: |
dadA |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in D-amino-acid dehydrogenase activity |
| Specific Function: |
Oxidative deamination of D-amino acids |
| Cellular Location: |
Cell inner membrane; Peripheral membrane protein |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
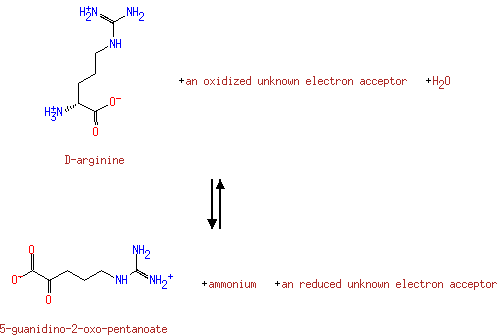
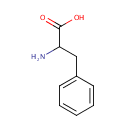
alpha-Amino acid | + |  | + | Acceptor | ↔ | 2-Oxo acid | + |  | + | Reduced acceptor |
| | |
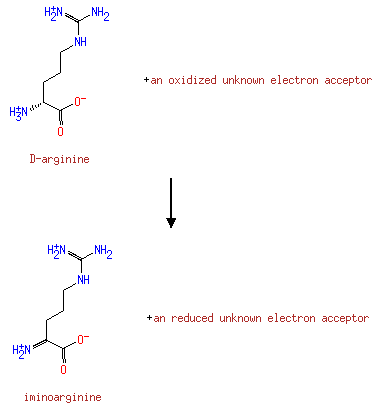
 | + |  | + | Acceptor | ↔ | 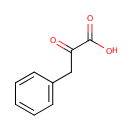 | + |  | + | Reduced acceptor |
| | |
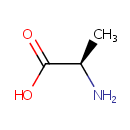
D-Amino acid | + |  | + | Acceptor | ↔ | 2-Oxo acid | + |  | + | Reduced acceptor |
| |
|
| SMPDB Reactions: |
| | |
 | + |  | + | cytochrome c nitrite reductase | ↔ |  | + |  | + | cytochrome c nitrite reductase |
| | |
 | + |  | + | an electron-transfer quinone | → |  | + | 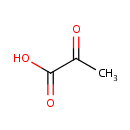 | + | electron-transfer quinol |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
| | |
A D-amino acid | + |  | + | acceptor | → | a 2-oxo acid | + |  | + | reduced acceptor |
| |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| catalytic activity | | oxidoreductase activity |
|
| Gene Properties |
| Locus tag: |
PA5304 |
| Strand: |
- |
| Entrez Gene ID: |
878057 |
| Accession: |
NP_253991.1 |
| GI: |
15600497 |
| Sequence start: |
5972178 |
| Sequence End: |
5973476 |
| Sequence Length: |
1298 |
| Gene Sequence: |
>PA5304
ATGCGAGTTCTGGTCCTTGGCAGCGGTGTCATCGGTACCGCCAGTGCGTATTACCTGGCCCGTGCCGGGTTCGAGGTGGTGGTGGTCGACCGTCAGGACGGTCCCGCGCTGGAAACCAGCTTCGCCAACGCCGGCCAGGTGTCTCCCGGCTACGCTTCGCCCTGGGCAGCCCCGGGCATTCCCCTGAAGGCCATGAAGTGGCTGCTGGAAAAGCACGCGCCGCTGGCCATCAAGCTCACCTCCGATCCCAGCCAGTACGCCTGGATGCTGCAGATGCTGCGCAACTGCACCGCCGAGCGCTACGCCGTGAACAAGGAGCGCATGGTCCGCCTGTCCGAGTACAGCCGCGATTGCCTCGACGAACTGCGCGCCGAGACCGGCATCGCCTACGAGGGCCGCACCCTCGGCACCACCCAACTGTTCCGCACCCAGGCGCAGCTGGACGCCGCCGGCAAGGACATCGCCGTGCTCGAGCGCTCCGGCGTGCCCTACGAGGTTCTCGACCGCGACGGCATCGCCCGCGTAGAGCCGGCTTTGGCCAAGGTCGCCGACAAGCTGGTCGGCGCCTTGCGCCTGCCCAACGACCAGACCGGCGACTGCCAGCTGTTCACCACCCGCCTGGCGGAAATGGCCAAGGGCCTGGGCGTGGAGTTCCGCTTCGGCCAGAACATCGAGCGCCTGGACTTCGCCGGCGACCGCATCAACGGCGTGCTGGTCAACGGCGAATTGCTCACCGCCGACCACTACGTGCTGGCCCTGGGCAGCTACTCGCCGCAACTGCTCAAGCCGCTGGGTATCAAGGCTCCGGTCTATCCGCTGAAGGGTTATTCGCTGACCGTGCCGATCACCAACCCGGAGATGGCGCCGACCTCGACCATCCTCGACGAGACCTACAAGGTGGCGATCACCCGCTTCGACCAGCGCATCCGCGTCGGCGGCATGGCGGAAATCGCCGGCTTCGACCTGTCGCTGAACCCGCGCCGCCGCGAGACCCTGGAAATGATCACCACCGACCTCTATCCCGAGGGCGGCGATATCAGCCAGGCGACCTTCTGGACCGGCCTGCGCCCGGCGACCCCGGATGGCACCCCGATCGTCGGCGCCACCCGCTACCGCAACCTGTTCCTCAATACCGGCCACGGCACCCTGGGTTGGACCATGGCCTGCGGGTCGGGTCGCTACCTGGCCGACCTGATGGCGAAGAAGCGCCCGCAGATCAGTACCGAAGGCCTGGATATTTCCCGCTACAGCAATTCCCCGGAGAACGCCAAGAATGCCCATCCAGCGCCAGCACACTAA |
| Protein Properties |
| Protein Residues: |
432 |
| Protein Molecular Weight: |
47.1 kDa |
| Protein Theoretical pI: |
7.49 |
| Hydropathicity (GRAVY score): |
-0.184 |
| Charge at pH 7 (predicted): |
1.09 |
| Protein Sequence: |
>PA5304
MRVLVLGSGVIGTASAYYLARAGFEVVVVDRQDGPALETSFANAGQVSPGYASPWAAPGIPLKAMKWLLEKHAPLAIKLTSDPSQYAWMLQMLRNCTAERYAVNKERMVRLSEYSRDCLDELRAETGIAYEGRTLGTTQLFRTQAQLDAAGKDIAVLERSGVPYEVLDRDGIARVEPALAKVADKLVGALRLPNDQTGDCQLFTTRLAEMAKGLGVEFRFGQNIERLDFAGDRINGVLVNGELLTADHYVLALGSYSPQLLKPLGIKAPVYPLKGYSLTVPITNPEMAPTSTILDETYKVAITRFDQRIRVGGMAEIAGFDLSLNPRRRETLEMITTDLYPEGGDISQATFWTGLRPATPDGTPIVGATRYRNLFLNTGHGTLGWTMACGSGRYLADLMAKKRPQISTEGLDISRYSNSPENAKNAHPAPAH |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA5304 and its homologs
|