| Identification |
| Name: |
Anthranilate synthase component II |
| Synonyms: |
- Glutamine amidotransferase
- Anthranilate phosphoribosyltransferase
|
| Gene Name: |
trpD |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in anthranilate phosphoribosyltransferase activity |
| Specific Function: |
Chorismate + L-glutamine = anthranilate + pyruvate + L-glutamate |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
| | |
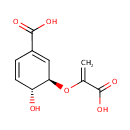
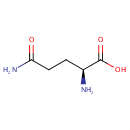
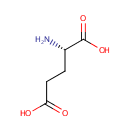
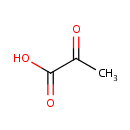
 | + |  | → | Pyrophosphate | + | N-(5-phosphoribosyl)-anthranilate | + |  |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| anthranilate phosphoribosyltransferase activity | | anthranilate synthase activity | | carbon-carbon lyase activity | | catalytic activity | | lyase activity | | oxo-acid-lyase activity | | transferase activity | | transferase activity, transferring glycosyl groups | | transferase activity, transferring pentosyl groups | | Process |
|---|
| biosynthetic process | | cellular amino acid and derivative metabolic process | | cellular amino acid derivative metabolic process | | cellular amino acid metabolic process | | cellular biogenic amine metabolic process | | cellular metabolic process | | glutamine family amino acid metabolic process | | glutamine metabolic process | | indolalkylamine metabolic process | | metabolic process | | tryptophan biosynthetic process | | tryptophan metabolic process |
|
| Gene Properties |
| Locus tag: |
PA0650 |
| Strand: |
+ |
| Entrez Gene ID: |
880665 |
| Accession: |
NP_249341.1 |
| GI: |
15595847 |
| Sequence start: |
704084 |
| Sequence End: |
705133 |
| Sequence Length: |
1049 |
| Gene Sequence: |
>PA0650
ATGGATATCAAGGGAGCCCTCAATCGCATCGTCAACCAGCTCGACCTGACCACCGAGGAAATGCAGGCGGTCATGCGCCAGATCATGACCGGGCAGTGCACCGACGCGCAGATCGGCGCCTTCCTGATGGGCATGCGGATGAAGAGCGAAACCATCGACGAGATCGTCGGCGCGGTGGCGGTGATGCGCGAACTGGCCGACGGCGTGCAGTTGCCTACGCTGAAGCATGTGGTCGACGTGGTCGGCACCGGCGGCGATGGCGCGAACATCTTCAACGTGTCCTCGGCGGCGTCCTTCGTGGTCGCCGCCGCTGGCGGCAAGGTCGCCAAACACGGTAACCGCGCGGTCTCCGGCAAGAGCGGCAGCGCCGACTTGCTGGAAGCCGCCGGCATCTACCTGGAGCTGACCTCCGAACAGGTGGCGCGTTGCATCGACACCGTCGGCGTCGGGTTCATGTTCGCCCAGGTCCACCACAAGGCGATGAAGTACGCCGCCGGTCCGCGCCGCGAGCTGGGCTTGCGGACTCTGTTCAACATGCTTGGCCCACTGACCAACCCGGCGGGAGTCAGGCACCAGGTGGTCGGGGTGTTCACCCAGGAACTGTGCAAGCCGCTGGCTGAAGTGCTCAAGCGTCTCGGCAGCGAGCATGTGCTGGTGGTGCATTCGCGCGACGGGCTGGACGAGTTCAGTCTGGCCGCGGCGACCCACATTGCCGAGTTGAAGGACGGCGAGGTACGCGAGTACGAAGTGCGTCCCGAGGACTTCGGGATCAAGAGCCAGACCCTGATGGGGCTGGAGGTCGACAGTCCGCAGGCCTCGCTGGAACTGATCCGCGACGCTTTGGGGCGGCGCAAGACCGAGGCTGGGCAGAAGGCCGCCGAGCTGATCGTGATGAATGCCGGCCCGGCACTGTACGCTGCCGATCTGGCGACCAGCCTGCACGAGGGCATTCAACTGGCCCACGATGCCCTGCACACCGGGCTGGCACGGGAGAAGATGGACGAACTGGTGGCCTTCACCGCCGTTTACAGAGAGGAGAACGCACAGTGA |
| Protein Properties |
| Protein Residues: |
349 |
| Protein Molecular Weight: |
37.4 kDa |
| Protein Theoretical pI: |
5.59 |
| Hydropathicity (GRAVY score): |
0.021 |
| Charge at pH 7 (predicted): |
-8.43 |
| Protein Sequence: |
>PA0650
MDIKGALNRIVNQLDLTTEEMQAVMRQIMTGQCTDAQIGAFLMGMRMKSETIDEIVGAVAVMRELADGVQLPTLKHVVDVVGTGGDGANIFNVSSAASFVVAAAGGKVAKHGNRAVSGKSGSADLLEAAGIYLELTSEQVARCIDTVGVGFMFAQVHHKAMKYAAGPRRELGLRTLFNMLGPLTNPAGVRHQVVGVFTQELCKPLAEVLKRLGSEHVLVVHSRDGLDEFSLAAATHIAELKDGEVREYEVRPEDFGIKSQTLMGLEVDSPQASLELIRDALGRRKTEAGQKAAELIVMNAGPALYAADLATSLHEGIQLAHDALHTGLAREKMDELVAFTAVYREENAQ |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA0650 and its homologs
|