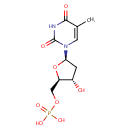
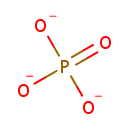
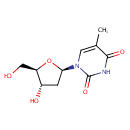
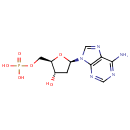
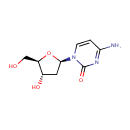
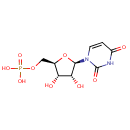
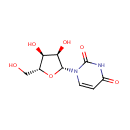
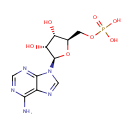
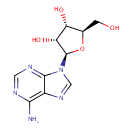
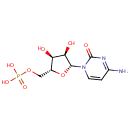
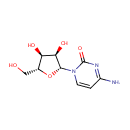
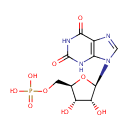
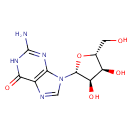
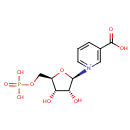
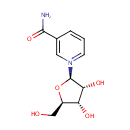
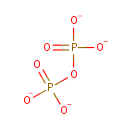
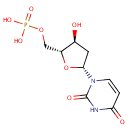
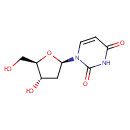
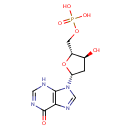
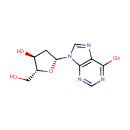
Multifunctional protein surE (PA3625)
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name: | Multifunctional protein surE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Name: | surE | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Enzyme Class: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General Function: | Involved in hydrolase activity | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Specific Function: | Nucleotidase with a broad substrate specificity as it can dephosphorylate various ribo- and deoxyribonucleoside 5'- monophosphates and ribonucleoside 3'-monophosphates with highest affinity to 3'-AMP. Also hydrolyzes polyphosphate (exopolyphosphatase activity) with the preference for short-chain- length substrates (P20-25). Might be involved in the regulation of dNTP and NTP pools, and in the turnover of 3'-mononucleotides produced by numerous intracellular RNases (T1, T2, and F) during the degradation of various RNAs. Also plays a significant physiological role in stress-response and is required for the survival of E.coli in stationary growth phase | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Location: | Cytoplasm (Potential) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Reactions: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Reactions: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PseudoCyc/BioCyc Reactions: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex Reactions: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Transports: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolites: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO Classification: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Locus tag: | PA3625 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Strand: | - | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Entrez Gene ID: | 880442 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession: | NP_252315.1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GI: | 15598821 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sequence start: | 4060625 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sequence End: | 4061374 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sequence Length: | 749 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Sequence: |
>PA3625 ATGCGCATACTGATTGCCAACGACGACGGGGTGACCGCACCCGGTATCGCCGCGCTTTACGACGCGCTGGCGGATCATGCCGATTGCGTAGTGATCGCCCCCGACCAGGACAAGAGCGGCGCCAGCAGTTCCCTGACGCTGGACCGCCCGCTGCACCCGCAGCGCCTGGACAACGGATTCATCAGCCTCAACGGCACGCCGACCGACTGCGTGCACCTGGGCCTGAACGGCTTGCTGGAGGAATTGCCGGACATGGTGGTCTCGGGTATCAACCTGGGCGCCAACCTCGGCGATGACGTCCTCTATTCCGGTACCGTGGCGGCGGCCATCGAAGGGCGCTTCCTGAAGGGGCCGGCCTTCGCCTTCTCGCTGGTCTCGCGGTTGACGGACAACCTGCCCACCGCCATGCACTTCGCCCGCCTGCTGGTCTCCGCCCACGAGCGCCTGGCCGTGCCGCCGCGTACCGTGCTCAACGTCAATATCCCGAACCTGCCGCTGGATCGCGTACGTGGTATCCAACTGACCCGCCTCGGGCACCGCGCCCGTGCCGCCGCACCGGTGAAGGTGGTCAATCCGCGCGGCAAGGAAGGCTACTGGATCGCCGCGGCCGGCGATGCCGAGGATGGCGGGCCGGGGACGGATTTCCACGCGGTCATGCAAGGCTACGTGTCGATCACCCCGTTGCAGCTGGACCGCACCTTCCACGAAGCCTTCGGCGGCCTCGACGAGTGGCTGGGAGGACTGACGTGA | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Residues: | 249 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Molecular Weight: | 26.4 kDa | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Theoretical pI: | 5.24 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Hydropathicity (GRAVY score): | 0.073 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge at pH 7 (predicted): | -7.14 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Sequence: |
>PA3625 MRILIANDDGVTAPGIAALYDALADHADCVVIAPDQDKSGASSSLTLDRPLHPQRLDNGFISLNGTPTDCVHLGLNGLLEELPDMVVSGINLGANLGDDVLYSGTVAAAIEGRFLKGPAFAFSLVSRLTDNLPTAMHFARLLVSAHERLAVPPRTVLNVNIPNLPLDRVRGIQLTRLGHRARAAAPVKVVNPRGKEGYWIAAAGDAEDGGPGTDFHAVMQGYVSITPLQLDRTFHEAFGGLDEWLGGLT | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General Reference: | PaperBLAST - Find papers about PA3625 and its homologs | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||