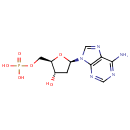
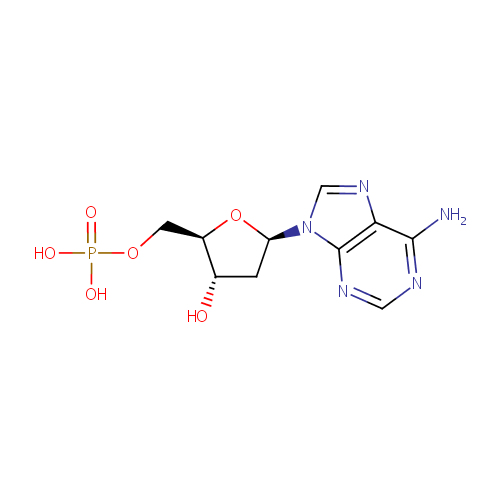
| References: |
- Avkin S, Adar S, Blander G, Livneh Z: Quantitative measurement of translesion replication in human cells: evidence for bypass of abasic sites by a replicative DNA polymerase. Proc Natl Acad Sci U S A. 2002 Mar 19;99(6):3764-9. Epub 2002 Mar 12. Pubmed: 11891323
- Bennett, B. D., Kimball, E. H., Gao, M., Osterhout, R., Van Dien, S. J., Rabinowitz, J. D. (2009). "Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli." Nat Chem Biol 5:593-599. Pubmed: 19561621
- Chen XR, Li GM, Wang JR, Chen JJ: [Portal hemodynamics in patients with different syndromes of cirrhosis] Zhong Xi Yi Jie He Xue Bao. 2004 May;2(3):178-81. Pubmed: 15339437
- Duarte V, Muller JG, Burrows CJ: Insertion of dGMP and dAMP during in vitro DNA synthesis opposite an oxidized form of 7,8-dihydro-8-oxoguanine. Nucleic Acids Res. 1999 Jan 15;27(2):496-502. Pubmed: 9862971
- Hashimoto K, Tominaga Y, Nakabeppu Y, Moriya M: Futile short-patch DNA base excision repair of adenine:8-oxoguanine mispair. Nucleic Acids Res. 2004 Nov 5;32(19):5928-34. Print 2004. Pubmed: 15531653
- Hohenester E, Hutchinson WL, Pepys MB, Wood SP: Crystal structure of a decameric complex of human serum amyloid P component with bound dAMP. J Mol Biol. 1997 Jun 20;269(4):570-8. Pubmed: 9217261
- Ishii, N., Nakahigashi, K., Baba, T., Robert, M., Soga, T., Kanai, A., Hirasawa, T., Naba, M., Hirai, K., Hoque, A., Ho, P. Y., Kakazu, Y., Sugawara, K., Igarashi, S., Harada, S., Masuda, T., Sugiyama, N., Togashi, T., Hasegawa, M., Takai, Y., Yugi, K., Arakawa, K., Iwata, N., Toya, Y., Nakayama, Y., Nishioka, T., Shimizu, K., Mori, H., Tomita, M. (2007). "Multiple high-throughput analyses monitor the response of E. coli to perturbations." Science 316:593-597. Pubmed: 17379776
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Levine RL, Yang IY, Hossain M, Pandya GA, Grollman AP, Moriya M: Mutagenesis induced by a single 1,N6-ethenodeoxyadenosine adduct in human cells. Cancer Res. 2000 Aug 1;60(15):4098-104. Pubmed: 10945616
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
- Zhang Q, Zhang WT, Wei JJ, Wang XB, Liu P: [Combined use of factor analysis and cluster analysis in classification of traditional Chinese medical syndromes in patients with posthepatitic cirrhosis] Zhong Xi Yi Jie He Xue Bao. 2005 Jan;3(1):14-8. Pubmed: 15644152
- Zhong H, Zang KT: Therapeutic approaches for chronic gastralgia based on differentiation of symptoms and signs. Di Yi Jun Yi Da Xue Xue Bao. 2002 Jul;22(7):639-40. Pubmed: 12376299
|
|---|