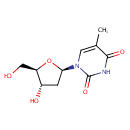
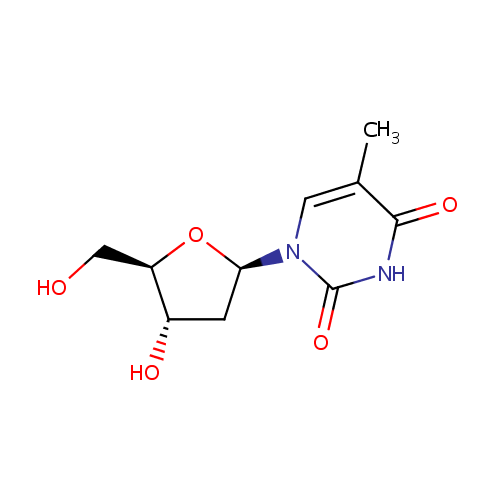
| References: |
- Abelson HT, Fosburg MT, Beardsley GP, Goorin AM, Gorka C, Link M, Link D: Methotrexate-induced renal impairment: clinical studies and rescue from systemic toxicity with high-dose leucovorin and thymidine. J Clin Oncol. 1983 Mar;1(3):208-16. Pubmed: 6607976
- Ashkenazi S, Cleary KR, Pickering LK, Murray BE, Cleary TG: The association of Shiga toxin and other cytotoxins with the neurologic manifestations of shigellosis. J Infect Dis. 1990 May;161(5):961-5. Pubmed: 2324546
- Eells JT, Spector R: Purine and pyrimidine base and nucleoside concentrations in human cerebrospinal fluid and plasma. Neurochem Res. 1983 Nov;8(11):1451-7. Pubmed: 6656991
- Grinlinton FM, Skinner MA, Birchall NM, Tan PL: Gamma delta + T cells from patients with psoriatic and rheumatoid arthritis respond to streptococcal antigen. J Rheumatol. 1993 Jun;20(6):982-7. Pubmed: 8350335
- Isoda K, Kim H, Hamamoto Y: A study on the mesothelial cell kinetics in pleural effusions by DNA cytophotometry and autoradiography with tritiated thymidine. Acta Pathol Jpn. 1984 Jul;34(4):775-83. Pubmed: 6485796
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Robins HI, Tutsch K, Katschinski DM, Jacobson E, Mehta M, Olsen M, Cohen JD, Tiggelaar CL, Arzoomanian RZ, Alberti D, Feierabend C, Wilding G: Phase I trial of intravenous thymidine and carboplatin in patients with advanced cancer. J Clin Oncol. 1999 Sep;17(9):2922-31. Pubmed: 10561372
- Schill WB, Miska W: Possible effects of the kallikrein-kinin system on male reproductive functions. Andrologia. 1992 Mar-Apr;24(2):69-75. Pubmed: 1318646
- Schulz CA, Mehta MP, Badie B, McGinn CJ, Robins HI, Hayes L, Chappell R, Volkman J, Binger K, Arzoomanian R, Simon K, Alberti D, Feierabend C, Tutsch KD, Kunugi KA, Wilding G, Kinsella TJ: Continuous 28-day iododeoxyuridine infusion and hyperfractionated accelerated radiotherapy for malignant glioma: a phase I clinical study. Int J Radiat Oncol Biol Phys. 2004 Jul 15;59(4):1107-15. Pubmed: 15234045
- Svendsen LB, Stener Jorgensen F, Hart Hansen O, Johansen A, Horn T, Larsen JK: Influence of the prostaglandin E1 analogue Rioprostil on the human gastric mucosa. Digestion. 1987;37(1):29-34. Pubmed: 3111919
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
- Vogel W, Schempp W, Sigwarth I: Comparison of thymidine, fluorodeoxyuridine, hydroxyurea, and methotrexate blocking at the G1/S phase transition of the cell cycle, studied by replication patterns. Hum Genet. 1978 Dec 18;45(2):193-8. Pubmed: 153886
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
- Zhao R, Zhang S, Hanscom M, Chattopadhyay S, Goldman ID: Loss of reduced folate carrier function and folate depletion result in enhanced pemetrexed inhibition of purine synthesis. Clin Cancer Res. 2005 Feb 1;11(3):1294-301. Pubmed: 15709201
|
|---|