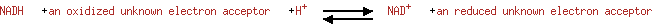| Identification |
| Name: |
NADH-quinone oxidoreductase subunit B |
| Synonyms: |
- NADH dehydrogenase I subunit B
- NDH-1 subunit B
- NUO2
|
| Gene Name: |
nuoB |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in NADH dehydrogenase (ubiquinone) activity |
| Specific Function: |
NDH-1 shuttles electrons from NADH, via FMN and iron- sulfur (Fe-S) centers, to quinones in the respiratory chain. The immediate electron acceptor for the enzyme in this species is ubiquinone. Couples the redox reaction to proton translocation (for every two electrons transferred, four hydrogen ions are translocated across the cytoplasmic membrane), and thus conserves the redox energy in a proton gradient |
| Cellular Location: |
Cell inner membrane; Peripheral membrane protein; Cytoplasmic side |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
 | + | Acceptor | ↔ | 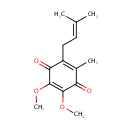 | + | Reduced acceptor |
| | | |
|
| SMPDB Reactions: |
| | |
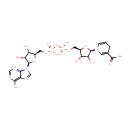 | + | 4  | + | 2  | + | menaquinone-8 | ? | 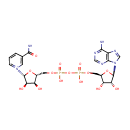 | + |  | + |  | + | Electron | + | 4  |
| | |
"a menaquinone" | + | 4  | + |  | → | "a menaquinol" | + |  |
| | |
4  | + |  | + | "a menaquinone" | → |  | + | "a menaquinol" |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| 4 iron, 4 sulfur cluster binding | | binding | | catalytic activity | | cofactor binding | | iron-sulfur cluster binding | | metal cluster binding | | NADH dehydrogenase (quinone) activity | | NADH dehydrogenase (ubiquinone) activity | | NADH dehydrogenase activity | | oxidoreductase activity | | oxidoreductase activity, acting on NADH or NADPH | | quinone binding | | Process |
|---|
| metabolic process | | oxidation reduction |
|
| Gene Properties |
| Locus tag: |
PA2638 |
| Strand: |
+ |
| Entrez Gene ID: |
882345 |
| Accession: |
NP_251328.1 |
| GI: |
15597834 |
| Sequence start: |
2983205 |
| Sequence End: |
2983882 |
| Sequence Length: |
677 |
| Gene Sequence: |
>PA2638
ATGCAATACAAACTTACCCGGATCGATCCCGATGCGGCCAACGACCAATACCCGATAGGCGAACGGGAAACCGTAGCCGACCCGCTGGTCGAGGGCCAGGTTCACAAGAACATCTTCATGGGCAAGCTCGAGGATGTGCTGAACTCCACCGTCAACTGGGGTCGCAAGAACTCGCTCTGGCCCTATAACTTCGGGCTGTCGTGCTGCTACGTGGAGATGACCACCGCCTTCACCGCGCCGCACGATATCGCCCGCTTCGGCGCCGAAGTGATCCGGGCGTCGCCGCGCCAGGCCGACTTCATGGTCATCGCCGGCACCTGCTTCATCAAGATGGCCCCGGTCATCCAGCGCCTCTACGAGCAGATGCTCGAACCCAAGTGGGTCATCTCCATGGGCTCGTGCGCCAATTCCGGCGGCATGTACGACATCTATTCGGTGGTCCAGGGGGTCGACAAGTTCCTCCCTGTGGACGTCTACATCCCCGGCTGCCCGCCCCGTCCGGAGGCGTTCCTGCAAGGCCTGATGCTGTTGCAGGAATCCATCGGCCAGGAGCGCCGCCCGCTGTCCTGGGTCGTTGGCGACCAGGGGGTCTACCGCGCCGAAATGCCGGCCCAGAAAGACCTGAAACGCGAGCAGCGCATCCAGGTGACCAACCTGCGCAGCCCCGACGAAGTGTGA |
| Protein Properties |
| Protein Residues: |
225 |
| Protein Molecular Weight: |
25.4 kDa |
| Protein Theoretical pI: |
5.1 |
| Hydropathicity (GRAVY score): |
-0.238 |
| Charge at pH 7 (predicted): |
-3.68 |
| Protein Sequence: |
>PA2638
MQYKLTRIDPDAANDQYPIGERETVADPLVEGQVHKNIFMGKLEDVLNSTVNWGRKNSLWPYNFGLSCCYVEMTTAFTAPHDIARFGAEVIRASPRQADFMVIAGTCFIKMAPVIQRLYEQMLEPKWVISMGSCANSGGMYDIYSVVQGVDKFLPVDVYIPGCPPRPEAFLQGLMLLQESIGQERRPLSWVVGDQGVYRAEMPAQKDLKREQRIQVTNLRSPDEV |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA2638 and its homologs
|