NADH (PAMDB110540)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB110540 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
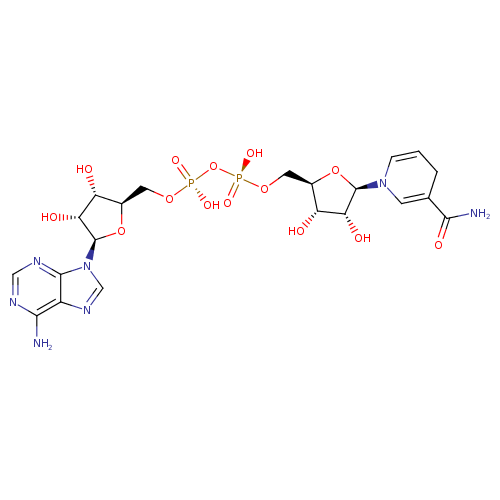
| Name: | NADH | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Dianion of NADH arising from deprotonation of the two diphosphate OH groups; major species at pH 7.3. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C21H27N7O14P2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 663.43 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 665.1247716967 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | BOPGDPNILDQYTO-NNYOXOHSSA-L | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C21H29N7O14P2/c22-17-12-19(25-7-24-17)28(8-26-12)21-16(32)14(30)11(41-21)6-39-44(36,37)42-43(34,35)38-5-10-13(29)15(31)20(40-10)27-3-1-2-9(4-27)18(23)33/h1,3-4,7-8,10-11,13-16,20-21,29-32H,2,5-6H2,(H2,23,33)(H,34,35)(H,36,37)(H2,22,24,25)/p-2/t10-,11-,13-,14-,15-,16-,20-,21-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 58-68-4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | adenosine 5'- | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | NADH | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | C1(=C(CC=CN1C5(OC(COP(=O)([O-])OP(=O)([O-])OCC2(OC(C(O)C(O)2)N4(C=NC3(C(N)=NC=NC=34))))C(O)C(O)5))C(N)=O) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of chemical entities known as (5'->5')-dinucleotides. These are dinucleotides where the two bases are connected via a (5'->5')-phosphodiester linkage. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Chemical entities | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Nucleosides, nucleotides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | (5'->5')-dinucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | (5'->5')-dinucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 140.0 - 142.0 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | 3,4-dihydroxyphenylglycol + NAD+ → 3,4-dihydroxyphenylglycolaldehyde + NADH + Hydrogen ion 3-[(3aS,4S,5R,7aS)-5-hydroxy-7a-methyl-1-oxo-octahydro-1H-inden-4-yl]-3-hydroxypropanoyl-CoA + NAD+ → 3-[(3aS,4S,5R,7aS)-5-hydroxy-7a-methyl-1-oxo-octahydro-1H-inden-4-yl]-3-oxopropanoyl-CoA + NADH + Hydrogen ion (S)-1-pyrroline-5-carboxylate + NAD+ + Water → Hydrogen ion + L-glutamate + NADH 4-phospho-hydroxy-L-threonine + NAD+ → 3-amino-1-hydroxyacetone 1-phosphate + Carbon dioxide + NADH methionol + NAD+ → 3-methylthiopropanal + NADH + Hydrogen ion 4-tyrosol + NAD+ → Hydrogen ion + (4-hydroxyphenyl)acetaldehyde + NADH 5,6-dihydrothymine + NAD+ → Thymine + NADH + Hydrogen ion (S)-3-hydroxyoctanoyl-CoA + NAD+ → 3-oxooctanoyl-CoA + NADH + Hydrogen ion (R,R)-2,3-butanediol + NAD+ → (R)-acetoin + NADH + Hydrogen ion NAD+ → NADH + Hydrogen ion D-glycerate + NAD+ → Hydrogen ion + tartronate semialdehyde + NADH formate + NAD+ → Carbon dioxide + NADH NADH + Water → Hydrogen ion + NMNH + AMP isobutanol + NAD+ → Hydrogen ion + isobutanal + NADH 3-hydroxypimeloyl-CoA + NAD+ → Hydrogen ion + NADH + 3-oxopimeloyl-CoA Ammonium + Water + NAD+ → Nitrite + NADH + Hydrogen ion benzaldehyde + Water + NAD+ → benzoate + NADH + Hydrogen ion 5,10-methylenetetrahydropteroyl mono-L-glutamate + NAD+ → 5,10-methenyltetrahydrofolate + NADH UDP-N-acetyl-α-D-mannosamine + NAD+ + Water → UDP-N-acetyl-α-D-mannosaminouronate + NADH + Hydrogen ion More...(S)-dihydroorotate + NAD+ → Hydrogen ion + orotate + NADH L-lactaldehyde + NAD+ + Water → (S)-lactate + NADH + Hydrogen ion 3-hydroxy-5-trans-dodecenoyl-CoA + NAD+ → 5-trans-3-oxo-dodecenoyl-CoA + NADH + Hydrogen ion 3-ethylmalate + NAD+ → 2-oxovalerate + Carbon dioxide + NADH UDP-α-D-glucuronate + NAD+ → UDP-4-Keto-pyranose + Carbon dioxide + NADH 4-acetamidobutanal + Water + NAD+ → 4-acetamidobutanoate + NADH + Hydrogen ion 3-oxopropanoate + coenzyme A + NAD+ → acetyl-CoA + Carbon dioxide + NADH L-glutamate + Water + NAD+ → alpha-Ketoglutarate + Ammonium + NADH + Hydrogen ion 3-hydroxy- 5-cis, 7-trans-tetradecadienoyl-CoA + NAD+ → 5-cis, 7-trans-3-oxo-tetradecadienoyl-CoA + NADH + Hydrogen ion 2-phenylethanol + NAD+ → Hydrogen ion + phenylacetaldehyde + NADH n-propanol + NAD+ → Propanal + NADH + Hydrogen ion 2-Keto-3-methyl-valerate + NAD+ + coenzyme A → 2-methylbutanoyl-CoA + NADH + Carbon dioxide UDP-N-acetyl-α-D-glucosamine + NAD+ + Water → UDP-N-acetyl-α-D-glucosaminouronate + NADH + Hydrogen ion D-threo-isocitrate + NAD+ → alpha-Ketoglutarate + Carbon dioxide + NADH 3-hydroxy, 4-trans-undecenoyl-CoA + NAD+ → 4-trans-3-oxo-undecenoyl-CoA + NADH + Hydrogen ion indole-3-glycol + NAD+ → indole-3-glycol aldehyde + NADH + Hydrogen ion 6-hydroxycyclohex-1-ene-1-carbonyl-CoA + NAD+ → Hydrogen ion + 6-oxocyclohex-1-ene-1-carbonyl-CoA + NADH L-3-HYDROXYACYL-COA + NAD+ → 3-KETOACYL-COA + NADH + Hydrogen ion Secondary-Alcohols + NAD+ → LONG-CHAIN-KETONE + NADH + Hydrogen ion Acetaldehyde + Water + NAD+ → acetate + NADH + Hydrogen ion NAD+ + 2-methyl-3-hydroxybutyryl-CoA → Hydrogen ion + NADH + 2-methylacetoacetyl-CoA OPC8-3-hydroxyacyl-CoA + NAD+ → OPC8-3-ketoacyl-CoA + NADH + Hydrogen ion 3-hydroxy, 6-trans-tridecenoyl-CoA + NAD+ → 6-trans-3-oxo-tridecenoyl-CoA + NADH + Hydrogen ion OPC6-3-hydroxyacyl-CoA + NAD+ → OPC6-3-ketoacyl-CoA + NADH + Hydrogen ion Propanal + coenzyme A + NAD+ → propanoyl-CoA + NADH + Hydrogen ion 2-methylbutanol + NAD+ → Hydrogen ion + 2-methylbutanal + NADH precorrin-2 + NAD+ → Hydrogen ion + sirohydrochlorin + NADH OPC4-3-hydroxyacyl-CoA + NAD+ → OPC4-3-ketoacyl-CoA + NADH + Hydrogen ion 3-hydroxy, 6-cis-tridecenoyl-CoA + NAD+ → 6-cis, 3-oxo-tridecenoyl-CoA + NADH + Hydrogen ion androsta-1,4-diene-3,17-dione + Oxygen + NADH + Hydrogen ion → 9α-hydroxyandrosta-1,4-diene-3,17-dione + NAD+ + Water 3-(4-hydroxyphenyl)-3-hydroxy-propanoyl-CoA + NAD+ → 4-hydroxybenzoyl-acetyl-CoA + NADH + Hydrogen ion (S)-3-hydroxyhexadecanoyl-CoA + NAD+ → 3-oxo-palmitoyl-CoA + NADH + Hydrogen ion 5-hydroxytryptophol + NAD+ → 5-hydroxyindole acetaldehyde + NADH + Hydrogen ion phytol + NAD+ → phytenal + NADH + Hydrogen ion phytenal + NAD+ + Water → phytenate + NADH + Hydrogen ion 3,5-cyclohexadiene-1,2-diol-1-carboxylate + NAD+ → catechol + Carbon dioxide + NADH 3-hydroxy-5-cis-tetradecenoyl-CoA + NAD+ → 3-keto-5-cis-tetradecenoyl-CoA + NADH + Hydrogen ion vanillin + Water + NAD+ → vanillate + NADH + Hydrogen ion Long-Chain-3S-Hydroxyacyl-CoAs + NAD+ → Long-Chain-oxoacyl-CoAs + NADH + Hydrogen ion 2-Me-Branched-234-Sat-FALD + NAD+ + Water → 2-Me-Branched-234-Sat-FA + NADH + Hydrogen ion 3-methoxy-4-hydroxyphenylglycol + NAD+ → 3-methoxy-4-hydroxyphenylglycolaldehyde + NADH + Hydrogen ion Odd-Straight-Chain-234-Sat-FALD + NAD+ + Water → Odd-Straight-Chain-234-Sat-FA + NADH + Hydrogen ion NAD+ + D-mannitol → aldehydo-D-mannose + Hydrogen ion + NADH n-butanol + NAD+ → Butanal + NADH + Hydrogen ion NADP+ + NADH + Hydrogen ion → NADPH + NAD+ + Hydrogen ion NADH + Hydrogen ion + Ubiquinones + NA+ → NAD+ + Ubiquinols + NA+ D-3-HYDROXYACYL-COA + NAD+ → 3-KETOACYL-COA + NADH + Hydrogen ion NADH + Hydrogen ion + ETR-Quinones → NAD+ + ETR-Quinols sn-glycerol 3-phosphate + NAD+ → glycerone phosphate + NADH + Hydrogen ion (S)-3-hydroxyhexanoyl-CoA + NAD+ → 3-oxohexanoyl-CoA + NADH + Hydrogen ion Aldehydes + NAD+ + Water → Carboxylates + NADH + Hydrogen ion Ubiquinones + Hydrogen ion + NADH → Ubiquinols + NAD+ FMNH(2) + NAD+ → FMN + NADH + Hydrogen ion 3-hydroxyadipyl-CoA + NAD+ → Hydrogen ion + 3-oxoadipyl-CoA + NADH 3-methylbutanol + NAD+ → Hydrogen ion + 3-methylbutanal + NADH Menaquinone + NADH + Hydrogen ion → menadiol + NAD+ D-arabitol + NAD+ → D-xylulose + NADH + Hydrogen ion cobalt-precorrin-6B + NAD+ → cobalt-precorrin-6A + NADH 5,6-dihydrouracil + NAD+ → Uracil + NADH + Hydrogen ion vanillate + Oxygen + NADH + Hydrogen ion → protocatechuate + Formaldehyde + NAD+ + Water 24-hydroxy-3-oxocholest-4-en-26-oyl-CoA + NAD+ → 3,24-dioxocholest-4-en-26-oyl-CoA + NADH + Hydrogen ion salicylaldehyde + NAD+ + Water → salicylate + NADH + Hydrogen ion UDP-N-acetyl-α-D-glucosaminouronate + NAD+ → UDP-2-acetamido-2-deoxy-α-D-ribo-hex-3-uluronate + NADH + Hydrogen ion histidinol + NAD+ + Water → L-Histidine + NADH + Hydrogen ion | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Marek, Miroslav; Vrbova, Eva; Horakova, Irena; Musil, Petr; Kefurt, Karel. NADH manufacture with immobilized Candida formate dehydrogenase. Czech. (1992), 4 pp. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||