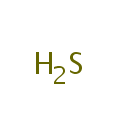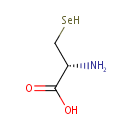| Identification |
| Name: |
Cysteine synthase A |
| Synonyms: |
- CSase A
- O-acetylserine (thiol)-lyase A
- OAS-TL A
- O-acetylserine sulfhydrylase A
- Sulfate starvation-induced protein 5
- SSI5
|
| Gene Name: |
cysK |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in cysteine biosynthetic process from serine |
| Specific Function: |
O(3)-acetyl-L-serine + H(2)S = L-cysteine + acetate |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
|
| KEGG Reactions: |
| | | | | | |
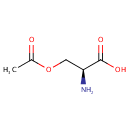 | + | 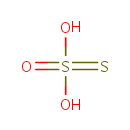 | + | Thioredoxin | + |  | ↔ | 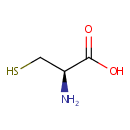 | + | 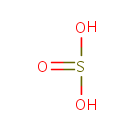 | + | Thioredoxin disulfide | + | 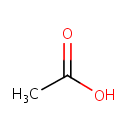 |
| |
|
| SMPDB Reactions: |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| binding | | carbon-oxygen lyase activity | | catalytic activity | | cofactor binding | | cysteine synthase activity | | lyase activity | | pyridoxal phosphate binding | | Process |
|---|
| cellular amino acid and derivative metabolic process | | cellular amino acid metabolic process | | cellular metabolic process | | cysteine biosynthetic process from serine | | L-serine metabolic process | | metabolic process | | serine family amino acid metabolic process |
|
| Gene Properties |
| Locus tag: |
PA2709 |
| Strand: |
+ |
| Entrez Gene ID: |
882613 |
| Accession: |
NP_251399.1 |
| GI: |
15597905 |
| Sequence start: |
3062939 |
| Sequence End: |
3063913 |
| Sequence Length: |
974 |
| Gene Sequence: |
>PA2709
ATGAGCCGCATCTTCGCAGACAACGCGCAGTCCATCGGCAACACGCCGCTGGTCCAGATCAACCGCATCGCCCCGCGCGGCGTCACCATCCTGGCCAAGATCGAAGGGCGCAACCCCGGCTATTCGGTGAAGTGCCGGATCGGCGCGAACATGATCTGGGATGCCGAAGCCAGCGGCAGACTGAAGTCCGGCATGACCCTGGTCGAGCCGACCTCCGGCAACACCGGCATCGGCCTCGCCTTCGTCGCCGCCGCACGCGGCTACAAACTGATCCTGACCATGCCGGCGTCGATGAGCCTGGAGCGACGCAAGGTGCTCAAGGCGCTGGGCGCGGAGCTGGTCCTGACCGAACCGGCGAAGGGCATGAAGGGTGCGATCCAGAAGGCCGAGGAACTGGTCGCGGGGGATCCCGGGAAGTACTTCATGCCGCAGCAGTTCGACAACCCGGCCAACCCGGCGATCCATGAGAAGACCACCGGCCCGGAGATCTGGAACGATACCGAGGGCGCGGTGGACGTGCTGGTCTCCGGGGTCGGCACCGGCGGCACCCTCACCGGCGTCTCGCGCTACATCAAGAACACCCGGGGCAAGCCGATCCTCGCCGTGGCGGTGGAGCCGGTGACCTCGCCGGTGATCAGCCAGACCCTCGCCGGCGAGGAGGTCAAGCCCGCCCCGCACAAGATCCAGGGCATCGGCGCCGGCTTCGTGCCGAAGAACCTCGACCTGTCGCTGGTCGACCGGGTCGAGAAGATCGGCGACGACGAAGCGAAGAACATGGCGCTGCGCCTGATGCAGGAAGAAGGCATCCTCTGCGGCATCTCCTCCGGCGCGGCGATGGCCGCCGCGGTGCGCCTGGCCGAGGAGCCGAACATGCAGGGCAAGACCATCGTGGTGATCCTCCCCGATTCCGGCGAGCGCTATCTTTCGAGCATGCTCTTCGACGGCCTGTTCAGCGAACAGGAACTGACCCAGTAA |
| Protein Properties |
| Protein Residues: |
324 |
| Protein Molecular Weight: |
34.3 kDa |
| Protein Theoretical pI: |
6.56 |
| Hydropathicity (GRAVY score): |
-0.037 |
| Charge at pH 7 (predicted): |
-0.57 |
| Protein Sequence: |
>PA2709
MSRIFADNAQSIGNTPLVQINRIAPRGVTILAKIEGRNPGYSVKCRIGANMIWDAEASGRLKSGMTLVEPTSGNTGIGLAFVAAARGYKLILTMPASMSLERRKVLKALGAELVLTEPAKGMKGAIQKAEELVAGDPGKYFMPQQFDNPANPAIHEKTTGPEIWNDTEGAVDVLVSGVGTGGTLTGVSRYIKNTRGKPILAVAVEPVTSPVISQTLAGEEVKPAPHKIQGIGAGFVPKNLDLSLVDRVEKIGDDEAKNMALRLMQEEGILCGISSGAAMAAAVRLAEEPNMQGKTIVVILPDSGERYLSSMLFDGLFSEQELTQ |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA2709 and its homologs
|