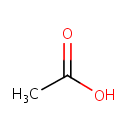
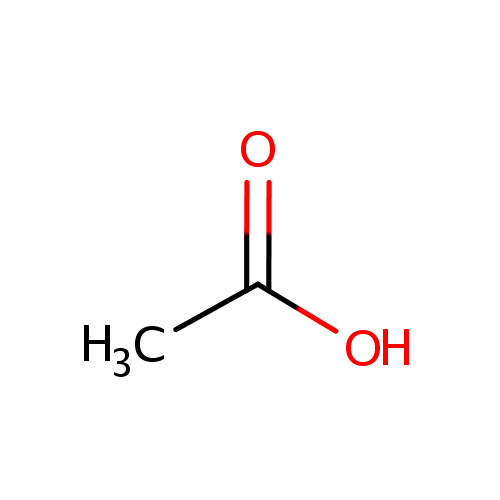
Acetic acid (PAMDB000012)
| Record Information | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000012 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Acetic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Acetic acid is one of the simplest carboxylic acids. The acetyl group, derived from acetic acid, is fundamental to the biochemistry of virtually all forms of life. When bound to coenzyme A it forms acetyl-CoA, which is central to the metabolism of carbohydrates and fats. However, the concentration of free acetic acid in cells is kept at a low level to avoid disrupting the control of the pH of the cell contents. Acetic acid is produced and excreted by certain bacteria, notably the Acetobacter genus and Clostridium acetobutylicum. These bacteria are found universally in foodstuffs, water, and soil, and acetic acid is produced naturally as fruits and some other foods spoil. (Wikipedia) Acetic acid, or more accurately, acetate, a derivative of acetic acid, can be produced by Pseudomonas aeruginosa through fermentation in glucose metabolism. (KEGG, PMID 18600996) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C2H4O2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 60.052 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 60.021129372 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | QTBSBXVTEAMEQO-UHFFFAOYSA-N | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C2H4O2/c1-2(3)4/h1H3,(H,3,4) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 64-19-7 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | acetic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | acetic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | CC(O)=O | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as carboxylic acids. These are compounds containing a carboxylic acid group with the formula -C(=O)OH. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Carboxylic acids | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Carboxylic acids | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Liquid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 16.6 °C | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Citric acid <> Acetic acid + Oxalacetic acid Acetoacetic acid + Acetyl-CoA > Acetoacetyl-CoA + Acetic acid Acetyl-CoA + Butyric acid > Acetic acid + Butyryl-CoA Acetyl-CoA + Hexanoate (N-C6:0) > Acetic acid + Hexanoyl-CoA O-Acetylserine + Hydrogen sulfide <> Acetic acid + L-Cysteine + Hydrogen ion Acetic acid + Adenosine triphosphate <> Acetylphosphate + ADP Water + UDP-3-O-(3-Hydroxymyristoyl)-N-acetylglucosamine <> Acetic acid + UDP-3-O-(3-Hydroxytetradecanoyl)-D-glucosamine N-Acetyl-D-Glucosamine 6-Phosphate + Water <> Acetic acid + Glucosamine 6-phosphate Water + Pyruvic acid + Ubiquinone-8 > Acetic acid + Carbon dioxide + Ubiquinol-8 Acetaldehyde + Water + NAD > Acetic acid +2 Hydrogen ion + NADH Acetaldehyde + Water + NADP > Acetic acid +2 Hydrogen ion + NADPH N-Acetylornithine + Water <> Acetic acid + Ornithine + L-Ornithine N-Acetyl-L-glutamate 5-semialdehyde + Water > Acetic acid + L-Glutamic-gamma-semialdehyde Acetic acid + Adenosine triphosphate + Coenzyme A <> Acetyl-CoA + Adenosine monophosphate + Pyrophosphate Acetylphosphate + Water <> Acetic acid + Phosphate Phosphonoacetate + Water <> Acetic acid + Phosphate N-Acetylornithine + Water <> Acetic acid + Ornithine O-Acetylserine + Hydrogen sulfide <> L-Cysteine + Acetic acid Butanoyl-CoA + Acetic acid <> Butyric acid + Acetyl-CoA O-Acetylserine + Thiosulfate <> Cysteine-S-sulfate + Acetic acid More...Pyruvic acid + Ubiquinone-1 + Water <> Acetic acid + Ubiquinol-8 + Carbon dioxide O-Acetylserine + Hydrogen selenide <> Selenocysteine + Acetic acid O-Acetylserine + Thiosulfate + Thioredoxin + Hydrogen ion <> L-Cysteine + Sulfite + Thioredoxin disulfide + Acetic acid 3-Hydroxy-5-oxohexanoate + Acetyl-CoA <> 3-Hydroxy-5-oxohexanoyl-CoA + Acetic acid N-Acetyl-L-citrulline + Water <> Acetic acid + Citrulline poly-β-1,6-N-acetyl-D-glucosamine + Water Hydrogen ion + partially N-deacetylated poly-β-1,6-N-acetyl-D-glucosamine + Acetic acid a 2,3,4-saturated fatty acyl CoA + Acetic acid <> a fatty acid + Acetyl-CoA Water + an acetic ester > an alcohol + Acetic acid + Hydrogen ion Pyruvic acid + Water + a ubiquinone > Carbon dioxide + a ubiquinol + Acetic acid Acetic acid + Carbon dioxide + Hydrogen ion <> Pyruvic acid + Water N-Acetyl-D-galactosamine 6-phosphate + Water > D-Galactosamine 6-phosphate + Acetic acid Acyl-CoA + Acetic acid > a fatty acid anion + Acetyl-CoA Acetyl-CoA + Citric acid > Acetic acid + (3S)-Citryl-CoA Adenosine triphosphate + Acetic acid + [citrate (pro-3S)-lyase](thiol form) > Adenosine monophosphate + Pyrophosphate + [citrate (pro-3S)-lyase](acetyl form) Pyruvic acid + Ubiquinone-10 + Water > Acetic acid + Carbon dioxide + Ubiquinol-1 Acyl-CoA + Acetic acid <> Fatty acid anion + Acetyl-CoA Adenosine triphosphate + Acetic acid + Citrate (pro-3S)-lyase (thiol form) <> Adenosine monophosphate + Pyrophosphate + Citrate (pro-3S)-lyase (acetyl form) Acetyl-CoA + Oxalic acid <> Acetic acid + Oxalyl-CoA Acetyl-CoA + 3-Hydroxy-5-oxohexanoate > Acetic acid + 3-Hydroxy-5-oxohexanoyl-CoA + 3-Hydroxy-5-oxohexanoyl-CoA N-Acetylornithine + Water > Acetic acid + L-Ornithine monochlorohydrate/ornithine N-Acetylornithine + Water > Ornithine + Acetic acid + Ornithine O-Acetylserine > Hydrogen ion + Acetic acid + L-Cysteine UDP-3-O-[(3R)-3-hydroxymyristoyl]-N-acetyl-α-D-glucosamine + Water > Acetic acid + UDP-3-O-(3-Hydroxytetradecanoyl)-D-glucosamine N-Acetyl-D-Glucosamine 6-Phosphate + Water + N-Acetyl-D-Glucosamine 6-Phosphate > Acetic acid + Glucosamine 6-phosphate O-Acetylserine + Thiosulfate + Thiosulfate > Cysteine-S-sulfate + Acetic acid | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Law, David John. Process for the preparation of carboxylic acids and/or derivatives thereof. PCT Int. Appl. (2007), 14pp. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in magnesium ion binding
- Specific function:
- Pyruvate + ferricytochrome b1 + H(2)O = acetate + CO(2) + ferrocytochrome b1
- Gene Name:
- poxB
- Locus Tag:
- PA5297
- Molecular weight:
- 62.3 kDa
Reactions
| Pyruvate + ubiquinone + H(2)O = acetate + CO(2) + ubiquinol. |
- General function:
- Involved in kinase activity
- Specific function:
- Involved in the activation of acetate to acetyl CoA and the secretion of acetate. During anaerobic growth of the organism, this enzyme is also involved in the synthesis of most of the ATP formed catabolically
- Gene Name:
- ackA
- Locus Tag:
- PA0836
- Molecular weight:
- 42.4 kDa
Reactions
| ATP + acetate = ADP + acetyl phosphate. |
| ATP + propanoate = ADP + propanoyl phosphate. |
- General function:
- Involved in UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase activity
- Specific function:
- The key enzyme in the biosynthesis of lipid A, a phosphorylated glycolipid that anchors the lipopolysaccharide to the outer membrane of the cell. Degraded by FtsH; when the activity of FtsH is reduced too much lipid A and not enough phospholipids are made (both pathways use the same precursor), which is lethal
- Gene Name:
- lpxC
- Locus Tag:
- PA4406
- Molecular weight:
- 33.4 kDa
Reactions
| UDP-3-O-(3-hydroxytetradecanoyl)-N-acetylglucosamine + H(2)O = UDP-3-O-(3-hydroxytetradecanoyl)-glucosamine + acetate. |
- General function:
- Involved in cysteine biosynthetic process from serine
- Specific function:
- O(3)-acetyl-L-serine + H(2)S = L-cysteine + acetate
- Gene Name:
- cysK
- Locus Tag:
- PA2709
- Molecular weight:
- 34.3 kDa
Reactions
| O(3)-acetyl-L-serine + H(2)S = L-cysteine + acetate. |
| 3-chloro-L-alanine + thioglycolate = S-carboxymethyl-L-cysteine + chloride. |
- General function:
- Involved in cysteine biosynthetic process from serine
- Specific function:
- Two cysteine synthase enzymes are found. Both catalyze the same reaction. Cysteine synthase B can also use thiosulfate in place of sulfide to give cysteine thiosulfonate as a product
- Gene Name:
- cysM
- Locus Tag:
- PA0932
- Molecular weight:
- 32.4 kDa
Reactions
| O(3)-acetyl-L-serine + H(2)S = L-cysteine + acetate. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Involved in the breakdown of putrescine. Was previously shown to have a weak but measurable ALDH enzyme activity that prefers NADP over NAD as coenzyme
- Gene Name:
- puuC
- Locus Tag:
- PA5312
- Molecular weight:
- 53.1 kDa
Reactions
| Gamma-glutamyl-gamma-aminobutyraldehyde + NAD(+) + H(2)O = gamma-glutamyl-gamma-aminobutyrate + NADH. |
| An aldehyde + NAD(P)(+) + H(2)O = a carboxylate + NAD(P)H. |
- General function:
- Involved in zinc ion binding
- Specific function:
- Displays a broad specificity and can also deacylate substrates such as acetylarginine, acetylhistidine or acetylglutamate semialdehyde
- Gene Name:
- argE
- Locus Tag:
- PA5206
- Molecular weight:
- 42.2 kDa
Reactions
| N(2)-acetyl-L-ornithine + H(2)O = acetate + L-ornithine. |
- General function:
- Involved in acetate-CoA ligase activity
- Specific function:
- Enables the cell to use acetate during aerobic growth to generate energy via the TCA cycle, and biosynthetic compounds via the glyoxylate shunt. Acetylates CheY, the response regulator involved in flagellar movement and chemotaxis
- Gene Name:
- acs
- Locus Tag:
- PA0887
- Molecular weight:
- 71.8 kDa
Reactions
| ATP + acetate + CoA = AMP + diphosphate + acetyl-CoA. |
- General function:
- Involved in ATP binding
- Specific function:
- Catalyzes two reactions:the first one is the production of beta-formyl glycinamide ribonucleotide (GAR) from formate, ATP and beta GAR; the second, a side reaction, is the production of acetyl phosphate and ADP from acetate and ATP
- Gene Name:
- purT
- Locus Tag:
- PA3751
- Molecular weight:
- 42.3 kDa
Reactions
| Formate + ATP + 5'-phospho-ribosylglycinamide = 5'-phosphoribosyl-N-formylglycinamide + ADP + diphosphate. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Catalyzes the NADP-dependent oxidation of diverse aldehydes such as chloroacetaldehyde, acetaldehyde, propionaldehyde, benzaldehyde, mafosfamide, 4- hydroperoxycyclophosphamide. Its preferred substrates are acetaldehyde and chloroacetaldehyde
- Gene Name:
- aldB
- Locus Tag:
- PA1984
- Molecular weight:
- 54.9 kDa
- General function:
- Not Available
- Specific function:
- Not Available
- Gene Name:
- yjdM
- Locus Tag:
- PA0128
- Molecular weight:
- 12.1 kDa
Transporters
- General function:
- Involved in transporter activity
- Specific function:
- Transports acetate. Also able to transport glycolate
- Gene Name:
- actP
- Locus Tag:
- PA3234
- Molecular weight:
- 58.7 kDa