| Identification |
| Name: |
Riboflavin synthase alpha chain |
| Synonyms: |
Not Available |
| Gene Name: |
ribE |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in riboflavin synthase activity |
| Specific Function: |
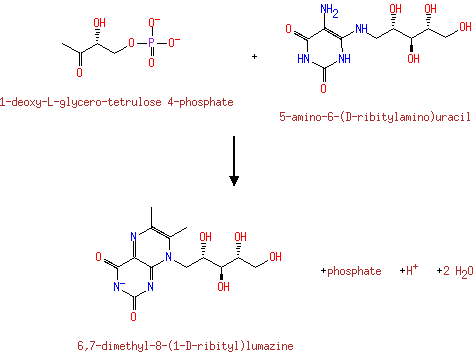
Riboflavin synthase is a bifunctional enzyme complex catalyzing the formation of riboflavin from 5-amino-6-(1'-D)- ribityl-amino-2,4(1H,3H)-pyrimidinedione and L-3,4-dihydrohy-2- butanone-4-phosphate via 6,7-dimethyl-8-lumazine. The alpha subunit catalyzes the dismutation of 6,7-dimethyl-8-lumazine to riboflavin and 5-amino-6-(1'-D)-ribityl-amino-2,4(1H,3H)- pyrimidinedione |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
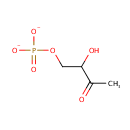
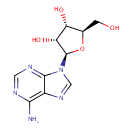
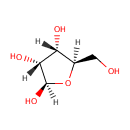
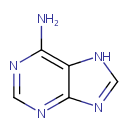
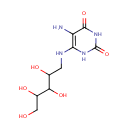 | + | 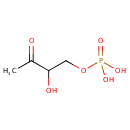 | → |  | + | 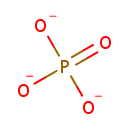 | + |  | + | 6,7-Dimethyl-8-(1-D-ribityl)lumazine | + | 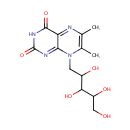 |
| | |
6,7-Dimethyl-8-(1-D-ribityl)lumazine | + |  | + |  | → | Riboflavin | + |  | + | 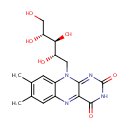 |
| | | | | |
|
| PseudoCyc/BioCyc Reactions: |
| | | | | | | 1-deoxy-L-glycero-tetrulose 4-phosphate + 5-amino-6-(D-ribitylamino)uracil → 6,7-dimethyl-8-(1-D-ribityl)lumazine + phosphate + H + + 2 H 2O ReactionCard |
|
| Complex Reactions: |
Not Available |
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| catalytic activity | | riboflavin synthase activity | | transferase activity | | transferase activity, transferring alkyl or aryl (other than methyl) groups | | Process |
|---|
| metabolic process | | nitrogen compound metabolic process | | riboflavin biosynthetic process | | riboflavin metabolic process |
|
| Gene Properties |
| Locus tag: |
PA4053 |
| Strand: |
- |
| Entrez Gene ID: |
879081 |
| Accession: |
NP_252742.1 |
| GI: |
15599248 |
| Sequence start: |
4534116 |
| Sequence End: |
4534592 |
| Sequence Length: |
476 |
| Gene Sequence: |
>PA4053
ATGACCCTGAAGACCATCGAAGGTACCTTCATTGCCCCCAAAGGCCGCTACGCCCTGGTGGTCGGCCGCTTCAACAGCTTCGTCGTGGAAAGCCTGGTCAGCGGCGCCGTCGACGCCCTGGTCCGCCACGGCGTGGCTGAAAGCGAGATCACCATCATCCGCGCCCCGGGTGCCTTCGAGATCCCGCTGGTGACCCAGAAGGTCGCCCAGCAAGGCGGCTTCGACGCCATCATCGCCCTCGGCGCGGTGATCCGTGGCGGCACCCCGCACTTCGAATACGTGGCTGGCGAGTGCACCAAGGGCCTGGCCCAGGTGTCCCTGCAGTTCGGCATCCCGGTCGCCTTCGGCGTCCTCACCGTCGACTCCATCGAGCAGGCCATCGAGCGCTCCGGCACCAAGGCCGGCAACAAGGGCGCCGAAGCCGCGCTGTCGGCCCTGGAAATGGTCAGCCTGCTGGCGCAGTTGGAGGCCAAGTGA |
| Protein Properties |
| Protein Residues: |
158 |
| Protein Molecular Weight: |
16.4 kDa |
| Protein Theoretical pI: |
5.75 |
| Hydropathicity (GRAVY score): |
0.449 |
| Charge at pH 7 (predicted): |
-1.56 |
| Protein Sequence: |
>PA4053
MTLKTIEGTFIAPKGRYALVVGRFNSFVVESLVSGAVDALVRHGVAESEITIIRAPGAFEIPLVTQKVAQQGGFDAIIALGAVIRGGTPHFEYVAGECTKGLAQVSLQFGIPVAFGVLTVDSIEQAIERSGTKAGNKGAEAALSALEMVSLLAQLEAK |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA4053 and its homologs
|