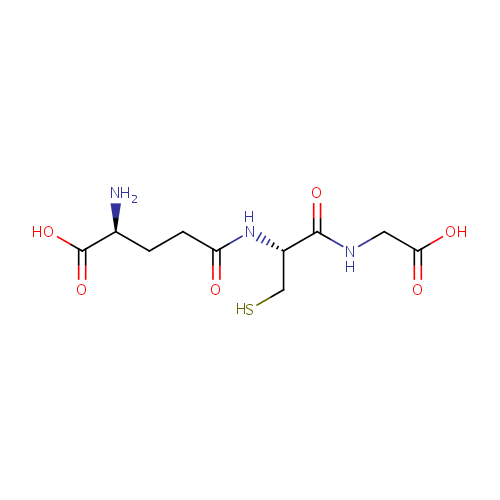
Glutathione (PAMDB000048)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000048 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Glutathione | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Glutathione (GSH) is a compound synthesized from cysteine. Like cysteine, glutathione contains the crucial thiol (-SH) group that makes it an effective antioxidant. There are virtually no living organisms on this planet-animal or plant whose cells don't contain some glutathione. Scientists have speculated that glutathione was essential to the very development of life on earth. Glutathione has many roles; in none does it act alone. It is a coenzyme in various enzymatic reactions. The most important of these are redox reactions, in which the thiol grouping on the cysteine portion of cell membranes protects against peroxidation; and conjugation reactions, in which glutathione binds with toxic chemicals in order to detoxify them. GSH is known as a substrate in both conjugation reactions and reduction reactions, catalyzed by glutathione S-transferase enzymes in the bacterial cytosol. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C10H17N3O6S | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 307.323 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 307.083805981 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | RWSXRVCMGQZWBV-WDSKDSINSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C10H17N3O6S/c11-5(10(18)19)1-2-7(14)13-6(4-20)9(17)12-3-8(15)16/h5-6,20H,1-4,11H2,(H,12,17)(H,13,14)(H,15,16)(H,18,19)/t5-,6-/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 70-18-8 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S)-2-amino-4-{[(1R)-1-[(carboxymethyl)carbamoyl]-2-sulfanylethyl]carbamoyl}butanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | glutathione | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | N[C@@H](CCC(=O)N[C@@H](CS)C(=O)NCC(O)=O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as gamma-glutamyl peptides. These are oligo- and polypeptides consisting of any C-terminal alpha peptide having a gamma-glutamyl residue attached at the N alpha-position. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Gamma-glutamyl peptides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 195 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Water + Glutathione > ADP + Glutathione + Hydrogen ion + Phosphate Adenosine triphosphate + Water + Glutathione > ADP + Glutathione + Hydrogen ion + Phosphate -->-->glutaredoxin + 2 Glutathione > glutaredoxin + Glutathione disulfide S-Formylglutathione + Water <> Formic acid + Glutathione + Hydrogen ion Arsenate + 2 Glutathione > Arsenite + Glutathione disulfide + Water Water + S-Lactoylglutathione > Glutathione + Hydrogen ion + D-Lactic acid periplasmic disulfide isomerase/thiol-disulphide oxidase (oxidized) + 2 Glutathione > periplasmic disulfide isomerase/thiol-disulphide oxidase (reduced) + Glutathione disulfide Glutathione + Pyruvaldehyde <> S-Lactoylglutathione 2 Glutathione + Hydrogen peroxide <> Glutathione disulfide +2 Water protein disulfide isomerase II (oxidized) + 2 Glutathione > protein disulfide isomerase II (reduced) + Glutathione disulfide Adenosine triphosphate + gamma-Glutamylcysteine + Glycine <> ADP + Glutathione + Hydrogen ion + Phosphate Adenosine triphosphate + Glutathione + Spermidine <> ADP + Glutathionylspermidine + Hydrogen ion + Phosphate Glutathionylspermidine + Water <> Glutathione + Spermidine Glutathione + Water > Cysteinylglycine + L-Glutamate Formaldehyde + Glutathione <> S-(Hydroxymethyl)glutathione 2 Glutathione + NAD <> Glutathione disulfide + NADH + Hydrogen ion 2 Glutathione + NADP <> Glutathione disulfide + NADPH + Hydrogen ion Adenosine triphosphate + gamma-Glutamylcysteine + Glycine <> ADP + Phosphate + Glutathione S-Formylglutathione + Water <> Formic acid + Glutathione Glutathione + L-Amino acid <> Cysteinylglycine + (5-L-Glutamyl)-L-amino acid S-Lactoylglutathione + Water <> Glutathione + D-Lactic acid More...Adenosine triphosphate + Glutathione + Spermidine <> ADP + Phosphate + Glutathionylspermidine RX + Glutathione <> Halide + R-S-Glutathione 1-Nitronaphthalene-7,8-oxide + Glutathione <> 1-Nitro-7-hydroxy-8-glutathionyl-7,8-dihydronaphthalene 1-Nitronaphthalene-7,8-oxide + Glutathione <> 1-Nitro-7-glutathionyl-8-hydroxy-7,8-dihydronaphthalene 1-Nitronaphthalene-5,6-oxide + Glutathione <> 1-Nitro-5-hydroxy-6-glutathionyl-5,6-dihydronaphthalene 1-Nitronaphthalene-5,6-oxide + Glutathione <> 1-Nitro-5-glutathionyl-6-hydroxy-5,6-dihydronaphthalene Bromobenzene-3,4-oxide + Glutathione <> 3,4-Dihydro-3-hydroxy-4-S-glutathionyl bromobenzene Bromobenzene-2,3-oxide + Glutathione <> 2,3-Dihydro-2-S-glutathionyl-3-hydroxy bromobenzene Benzo[a]pyrene-4,5-oxide + Glutathione <> 4,5-Dihydro-4-hydroxy-5-S-glutathionyl-benzo[a]pyrene Benzo[a]pyrene-7,8-diol + Glutathione <> 7,8-Dihydro-7-hydroxy-8-S-glutathionyl-benzo[a]pyrene + Water 2,2-Dichloroacetaldehyde + Glutathione <> S-(2,2-Dichloro-1-hydroxy)ethyl glutathione 2,2-Dichloroacetaldehyde + Glutathione <> S-(2-Chloroacetyl)glutathione + Hydrochloric acid 2-(S-Glutathionyl)acetyl chloride + Glutathione <> 2-(S-Glutathionyl)acetyl glutathione + Hydrochloric acid Trichloroethene + Glutathione <> S-(1,2-Dichlorovinyl)glutathione + Hydrochloric acid 1,2-Dibromoethane + Glutathione + Hydrogen ion <> Glutathione episulfonium ion +2 Hydrobromic acid Aflatoxin B1-exo-8,9-epoxide + Glutathione <> Aflatoxin B1exo-8,9-epoxide-GSH Selenite + Glutathione + Hydrogen ion > Selenodiglutathione + Glutathione disulfide + Water -->-->2-hydroxyethyldisulfide + Glutathione 2-mercaptoethanol + Glutathione disulfide bromoacetate + Glutathione Hydrogen ion + glutathione-S-acetate + Br<SUP>-</SUP> -->-->S-(2-hydroxyacyl)glutathione + Water > Glutathione + a 2-hydroxy carboxylate Adenosine triphosphate + gamma-Glutamylcysteine + Glycine > ADP + Inorganic phosphate + Glutathione 2 Glutathione + NADP > Glutathione disulfide + NADPH Glutathione + Spermidine + Adenosine triphosphate > Glutathionylspermidine + ADP + Inorganic phosphate RX + Glutathione > HX + R-S-glutathione RX + Glutathione <> Halide + R-S-Glutathione S-(2-Hydroxyacyl)glutathione + Water <> Glutathione + 2-Hydroxy carboxylate gamma-Glutamylcysteine + Glycine + Adenosine triphosphate > Hydrogen ion + Phosphate + Adenosine diphosphate + Glutathione + ADP Oxidized glutathione + Hydrogen ion + NADPH + Glutathione disulfide + NADPH > NADP +2 Glutathione Naphthalene epoxide + Glutathione + (1R,2S)-Naphthalene 1,2-oxide > (1R)-Glutathionyl-(2R)-hydroxy-1,2-dihydronaphthalene Naphthalene epoxide + Glutathione + (1R,2S)-Naphthalene 1,2-oxide > (1R)-Hydroxy-(2R)-glutathionyl-1,2-dihydronaphthalene Glutathione + Naphthalene epoxide + (1R,2S)-Naphthalene 1,2-oxide > (1S)-Hydroxy-(2S)-glutathionyl-1,2-dihydronaphthalene Glutathione + 2,2-Dichloroacetaldehyde > S-(Formylmethyl)glutathione | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in ATP binding
- Specific function:
- ATP + gamma-L-glutamyl-L-cysteine + glycine = ADP + phosphate + glutathione
- Gene Name:
- gshB
- Locus Tag:
- PA0407
- Molecular weight:
- 35.7 kDa
Reactions
| ATP + gamma-L-glutamyl-L-cysteine + glycine = ADP + phosphate + glutathione. |
- General function:
- Involved in oxidoreductase activity
- Specific function:
- Maintains high levels of reduced glutathione in the cytosol
- Gene Name:
- gor
- Locus Tag:
- PA2025
- Molecular weight:
- 49.2 kDa
Reactions
| 2 glutathione + NADP(+) = glutathione disulfide + NADPH. |
- General function:
- Involved in arsenate reductase (glutaredoxin) activity
- Specific function:
- Reduction of arsenate [As(V)] to arsenite [As(III)]. This protein expands the substrate specificity of ArsAB pump which can extrude arsenite and antimonite to allow for arsenate pumping and resistance
- Gene Name:
- arsC
- Locus Tag:
- PA2279
- Molecular weight:
- 16.6 kDa
Reactions
| Arsenate + glutaredoxin = arsenite + glutaredoxin disulfide + H(2)O. |
- General function:
- Involved in lactoylglutathione lyase activity
- Specific function:
- Catalyzes the conversion of hemimercaptal, formed from methylglyoxal and glutathione, to S-lactoylglutathione
- Gene Name:
- gloA
- Locus Tag:
- PA3524
- Molecular weight:
- 14.3 kDa
Reactions
| (R)-S-lactoylglutathione = glutathione + methylglyoxal. |
- General function:
- Involved in gamma-glutamyltransferase activity
- Specific function:
- (5-L-glutamyl)-peptide + an amino acid = peptide + 5-L-glutamyl amino acid
- Gene Name:
- ggt
- Locus Tag:
- PA1338
- Molecular weight:
- 59.9 kDa
Reactions
| A (5-L-glutamyl)-peptide + an amino acid = a peptide + a 5-L-glutamyl amino acid. |
| Glutathione + H(2)O = L-cysteinylglycine + L-glutamate. |
- General function:
- Involved in carboxylesterase activity
- Specific function:
- Serine hydrolase involved in the detoxification of formaldehyde. Hydrolyzes S-formylglutathione to glutathione and formate. Shows also esterase activity against alpha-naphthyl acetate, lactoylglutathione, palmitoyl-CoA and several pNP-esters of short chain fatty acids
- Gene Name:
- yeiG
- Locus Tag:
- PA3628
- Molecular weight:
- 31.2 kDa
Reactions
| S-formylglutathione + H(2)O = glutathione + formate. |
- General function:
- Posttranslational modification, protein turnover, chaperones
- Specific function:
- Involved in disulfide bond formation. DsbG and DsbC are part of a periplasmic reducing system that controls the level of cysteine sulfenylation, and provides reducing equivalents to rescue oxidatively damaged secreted proteins such as ErfK, YbiS and YnhG. Probably also functions as a disulfide isomerase with a narrower substrate specificity than DsbC. DsbG is maintained in a reduced state by DsbD. Displays chaperone activity in both redox states in vitro
- Gene Name:
- dsbG
- Locus Tag:
- PA2476
- Molecular weight:
- 28.1 kDa
- General function:
- Involved in electron carrier activity
- Specific function:
- Monothiol glutaredoxin involved in the biogenesis of iron-sulfur clusters (Probable)
- Gene Name:
- grxD
- Locus Tag:
- PA3533
- Molecular weight:
- 11.8 kDa
- General function:
- Involved in cell redox homeostasis
- Specific function:
- Acts as a disulfide isomerase, interacting with incorrectly folded proteins to correct non-native disulfide bonds. DsbG and DsbC are part of a periplasmic reducing system that controls the level of cysteine sulfenylation, and provides reducing equivalents to rescue oxidatively damaged secreted proteins. Acts by transferring its disulfide bond to other proteins and is reduced in the process. DsbC is reoxidized by DsbD
- Gene Name:
- dsbC
- Locus Tag:
- PA3737
- Molecular weight:
- 26.1 kDa
- General function:
- Involved in electron carrier activity
- Specific function:
- The disulfide bond functions as an electron carrier in the glutathione-dependent synthesis of deoxyribonucleotides by the enzyme ribonucleotide reductase. In addition, it is also involved in reducing some disulfides in a coupled system with glutathione reductase
- Gene Name:
- grxC
- Locus Tag:
- PA5129
- Molecular weight:
- 9.2 kDa
- General function:
- Not Available
- Specific function:
- Not Available
- Gene Name:
- yfcG
- Locus Tag:
- PA1033
- Molecular weight:
- 24.5 kDa