| Identification |
| Name: |
UDP-N-acetylglucosamine--N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase |
| Synonyms: |
- Undecaprenyl-PP-MurNAc-pentapeptide-UDPGlcNAc GlcNAc transferase
|
| Gene Name: |
murG |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in undecaprenyldiphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase activity |
| Specific Function: |
Cell wall formation. Catalyzes the transfer of a GlcNAc subunit on undecaprenyl-pyrophosphoryl-MurNAc-pentapeptide (lipid intermediate I) to form undecaprenyl-pyrophosphoryl-MurNAc- (pentapeptide)GlcNAc (lipid intermediate II) |
| Cellular Location: |
Cell inner membrane; Peripheral membrane protein |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
Undecaprenyl-diphospho-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimeloyl-D-alanyl-D-alanine | + | 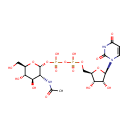 | + | 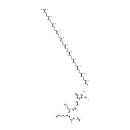 | → | Uridine 5'-diphosphate | + |  | + |  | + | 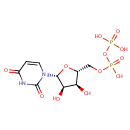 |
| | | |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
| PAMDB003414 | GlcNAc-(1->4)-Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-diphosphoundecaprenol | MetaboCard | | PAMDB001633 | Hydrogen ion | MetaboCard | | PAMDB003413 | lipid II(A) | MetaboCard | | PAMDB003420 | Mur2Ac(oyl-L-Ala-gamma-D-Glu-L-Lys-D-Ala-D-Ala)-diphosphoundecaprenol | MetaboCard | | PAMDB001020 | MurAc(oyl-L-Ala-D-gamma-Glu-L-Lys-D-Ala-D-Ala)-diphospho-undecaprenol | MetaboCard | | PAMDB001129 | N-Acetylmuramoyl-L-alanyl-D-glutamyl-L-lysyl-D-alanyl-D-alanine-diphosphoundecaprenyl-N-acetylglucosamine | MetaboCard | | PAMDB001100 | N-Acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimelyl-D-alanyl-D-alanine-diphosphoundecaprenol | MetaboCard | | PAMDB001056 | Undecaprenyl-diphospho-N-acetylmuramoyl-(N-acetylglucosamine)-L-alanyl-D-glutaminyl-meso-2,6-diaminopimeloyl-D-alanyl-D-alanine | MetaboCard | | PAMDB001058 | Undecaprenyl-diphospho-N-acetylmuramoyl-(N-acetylglucosamine)-L-alanyl-D-glutamyl-meso-2,6-diaminopimeloyl-D-alanyl-D-alanine | MetaboCard | | PAMDB001060 | Undecaprenyl-diphospho-N-acetylmuramoyl-(N-acetylglucosamine)-L-alanyl-gamma-D-glutamyl-L-lysyl-D-alanyl-D-alanine | MetaboCard | | PAMDB001062 | Undecaprenyl-diphospho-N-acetylmuramoyl-L-alanyl-D-glutamyl-meso-2,6-diaminopimeloyl-D-alanyl-D-alanine | MetaboCard | | PAMDB000123 | Uridine 5'-diphosphate | MetaboCard | | PAMDB000120 | Uridine diphosphate-N-acetylglucosamine | MetaboCard |
|
| GO Classification: |
| Function |
|---|
| acetylglucosaminyltransferase activity | | binding | | carbohydrate binding | | catalytic activity | | transferase activity | | transferase activity, transferring glycosyl groups | | transferase activity, transferring hexosyl groups | | UDP-glycosyltransferase activity | | undecaprenyldiphospho-muramoylpentapeptide beta-N-acetylglucosaminyltransferase activity | | Process |
|---|
| alcohol metabolic process | | amino sugar metabolic process | | carbohydrate metabolic process | | cellular lipid metabolic process | | lipid glycosylation | | lipid metabolic process | | lipid modification | | metabolic process | | monosaccharide metabolic process | | primary metabolic process | | small molecule metabolic process | | UDP-N-acetylgalactosamine biosynthetic process | | UDP-N-acetylgalactosamine metabolic process |
|
| Gene Properties |
| Locus tag: |
PA4412 |
| Strand: |
- |
| Entrez Gene ID: |
881264 |
| Accession: |
NP_253102.1 |
| GI: |
15599608 |
| Sequence start: |
4945068 |
| Sequence End: |
4946141 |
| Sequence Length: |
1073 |
| Gene Sequence: |
>PA4412
ATGAAAGGTAATGTCCTGATCATGGCGGGTGGCACCGGCGGACACGTGTTCCCGGCGCTCGCCTGTGCGCGGGAGTTCCAGGCGCGCGGCTATGCCGTGCACTGGCTGGGGACGCCGCGCGGCATCGAGAATGACCTGGTACCCAAGGCCGGCCTGCCGTTGCACCTGATCCAGGTCAGCGGGCTGCGCGGCAAGGGCCTGAAGTCGCTGGTCAAGGCGCCGCTGGAACTGCTCAAGTCGCTGTTCCAGGCGCTGCGGGTGATCCGCCAGCTGAGGCCGGTCTGCGTGCTCGGCCTGGGTGGCTATGTCACCGGCCCGGGCGGCCTGGCTGCGCGCCTCAACGGCGTGCCGCTGGTGATCCACGAGCAGAACGCCGTGGCCGGCACCGCCAACCGCAGCCTGGCGCCGATCGCCAGGCGCGTCTGCGAGGCATTCCCGGATACCTTCCCGGCCAGTGACAAGCGTCTGACCACCGGCAATCCGGTGCGCGGCGAGCTGTTCCTCGACGCGCACGCCCGCGCGCCGCTGACCGGCCGTCGGGTCAATCTGCTGGTGCTCGGCGGCAGCCTTGGCGCGGAACCGCTGAACAAGCTGTTGCCAGAGGCCCTGGCGCAGGTGCCGCTGGAAATCCGTCCGGCAATCCGCCATCAGGCCGGTCGTCAGCATGCCGAGATTACCGCCGAGCGTTATCGCACAGTGGCGGTCGAAGCGGACGTCGCGCCCTTCATCAGCGACATGGCCGCGGCCTATGCCTGGGCCGACCTGGTGATCTGTCGCGCCGGCGCGCTGACCGTCAGCGAGCTGACGGCGGCGGGATTGCCCGCCTTCCTGGTGCCGTTGCCTCACGCGATCGACGATCACCAGACCCGCAATGCCGAATTCCTGGTACGCAGCGGCGCCGGCCGCCTGCTGCCGCAAAAGTCTACCGGCGCGGCCGAACTGGCCGCGCAGCTGTCCGAGGTCCTGATGCATCCCGAAACCCTGCGCTCCATGGCAGACCAGGCACGCAGCCTGGCGAAACCCGAGGCTACCCGGACGGTGGTCGATGCCTGCCTGGAGGTGGCCCGTGGTTAA |
| Protein Properties |
| Protein Residues: |
357 |
| Protein Molecular Weight: |
37.8 kDa |
| Protein Theoretical pI: |
9.99 |
| Hydropathicity (GRAVY score): |
0.138 |
| Charge at pH 7 (predicted): |
11.25 |
| Protein Sequence: |
>PA4412
MKGNVLIMAGGTGGHVFPALACAREFQARGYAVHWLGTPRGIENDLVPKAGLPLHLIQVSGLRGKGLKSLVKAPLELLKSLFQALRVIRQLRPVCVLGLGGYVTGPGGLAARLNGVPLVIHEQNAVAGTANRSLAPIARRVCEAFPDTFPASDKRLTTGNPVRGELFLDAHARAPLTGRRVNLLVLGGSLGAEPLNKLLPEALAQVPLEIRPAIRHQAGRQHAEITAERYRTVAVEADVAPFISDMAAAYAWADLVICRAGALTVSELTAAGLPAFLVPLPHAIDDHQTRNAEFLVRSGAGRLLPQKSTGAAELAAQLSEVLMHPETLRSMADQARSLAKPEATRTVVDACLEVARG |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA4412 and its homologs
|