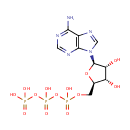
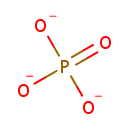
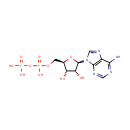
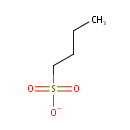
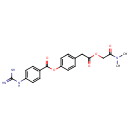
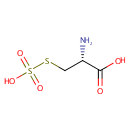
Putative aliphatic sulfonates transport permease protein ssuC (PA3443)
| Identification | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name: | Putative aliphatic sulfonates transport permease protein ssuC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Name: | ssuC | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Enzyme Class: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General Function: | Involved in transporter activity | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Specific Function: | Part of a binding-protein-dependent transport system for aliphatic sulfonates. Probably responsible for the translocation of the substrate across the membrane | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Location: | Cell inner membrane; Multi-pass membrane protein | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Pathways: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| KEGG Reactions: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMPDB Reactions: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| PseudoCyc/BioCyc Reactions: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Complex Reactions: | Not Available | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Transports: | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolites: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GO Classification: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Locus tag: | PA3443 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Strand: | - | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Entrez Gene ID: | 879101 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Accession: | NP_252133.1 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| GI: | 15598639 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sequence start: | 3848795 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sequence End: | 3849583 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sequence Length: | 788 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Gene Sequence: |
>PA3443 ATGAGCATTTCCAAACGACTGGCCGAGCGCCTGGCGCCCTGGGCGCTGCCGCTGGTCCTGCTGGCGGTGTGGCAGGCGGCGGTTGAGTTCGGCTGGCTGTCCAGCCGCATCCTCCCCGCGCCCAGCGCCGTACTCGAGGCCGGCTGGGCGCTGCTGCGCAGCGGCGAGATCTGGCAGCACCTGGCGATCAGCGGCTGGCGCGCGGCGATCGGCTTTCTCATCGGCGGCGCCATCGGCCTGGTGCTGGGCTTCGTCACCGGCCTGTCGAAGTGGGGCGAACGCTTCCTCGACAGTTCGGTGCAGATGATCCGCAACGTGCCGCACCTGGCGCTGATCCCGCTGGTGATCCTGTGGTTCGGCATCGACGAGTCGGCGAAGATCTTCCTGGTCGCCCTCGGCACGCTGTTCCCGATCTACCTGAACACCTACCACGGCATCCGCGGCATCGACCCCGGCCTGCTGGAGATGGCGCGCAGCTACGGGTTGTCCGGCTTCGCCCTGTTCCGCCAGGTGATCCTGCCCGGCGCGCTGCCGTCGATCCTGGTCGGGGTGCGCTTCGCCCTTGGCTTCATGTGGTTGACCCTGATCGTCGCCGAGACCATTTCCGCCAGCTCCGGCATCGGTTACCTGGCGATGAACGCCCGCGAGTTCCTGCAGACCGACGTGGTGGTCCTGGCGATCCTGCTCTATGCCGTCCTTGGCAAGCTCGCCGACCTCGCCGCCCGCGGCCTGGAACGCGTCTGGCTACGCTGGCACCCGGCCTACCAGGTCAAGGGAGGTGGCGCATGA | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Properties | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Residues: | 262 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Molecular Weight: | 28.5 kDa | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Theoretical pI: | 10.13 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Hydropathicity (GRAVY score): | 0.771 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge at pH 7 (predicted): | 5.95 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Protein Sequence: |
>PA3443 MSISKRLAERLAPWALPLVLLAVWQAAVEFGWLSSRILPAPSAVLEAGWALLRSGEIWQHLAISGWRAAIGFLIGGAIGLVLGFVTGLSKWGERFLDSSVQMIRNVPHLALIPLVILWFGIDESAKIFLVALGTLFPIYLNTYHGIRGIDPGLLEMARSYGLSGFALFRQVILPGALPSILVGVRFALGFMWLTLIVAETISASSGIGYLAMNAREFLQTDVVVLAILLYAVLGKLADLAARGLERVWLRWHPAYQVKGGGA | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| General Reference: | PaperBLAST - Find papers about PA3443 and its homologs | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||