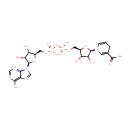
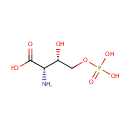| Identification |
| Name: |
Pyridoxine 5'-phosphate synthase |
| Synonyms: |
|
| Gene Name: |
pdxJ |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in catalytic activity |
| Specific Function: |
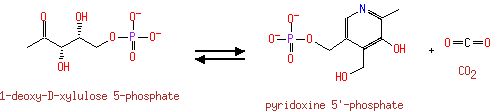
Catalyzes the complicated ring closure reaction between the two acyclic compounds 1-deoxy-D-xylulose-5-phosphate (DXP) and 3-amino-2-oxopropyl phosphate (1-amino-acetone-3-phosphate or AAP) to form pyridoxine 5'-phosphate (PNP) and inorganic phosphate |
| Cellular Location: |
Cytoplasm |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
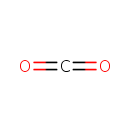
3-Amino-2-oxopropyl phosphate | + | 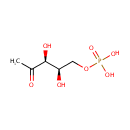 | + | 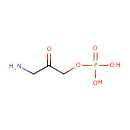 | ↔ | 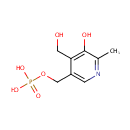 | + | 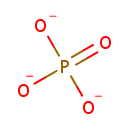 | + | 2  |
| |
|
| SMPDB Reactions: |
|
1-Deoxy-D-xylulose 5-phosphate | + | 2-Amino-3-phosphonopropionic acid | + |  | + | 2-Amino-3-phosphonopropionic acid | → |  | + |  | + |  | + | 2  |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Component |
|---|
| cell part | | cytoplasm | | intracellular part | | Function |
|---|
| catalytic activity | | Process |
|---|
| metabolic process | | pyridoxine biosynthetic process | | pyridoxine metabolic process | | small molecule metabolic process | | vitamin B6 metabolic process | | vitamin metabolic process | | water-soluble vitamin metabolic process |
|
| Gene Properties |
| Locus tag: |
PA0773 |
| Strand: |
+ |
| Entrez Gene ID: |
878131 |
| Accession: |
NP_249464.1 |
| GI: |
15595970 |
| Sequence start: |
841079 |
| Sequence End: |
841825 |
| Sequence Length: |
746 |
| Gene Sequence: |
>PA0773
GTGACTGAAGCCACTCGAATTCTGCTCGGCGTGAACATCGATCACGTCGCCACCCTGCGCCAGGCCCGGGGCACCCGCTATCCGGACCCGGTGAAGGCCGCCCTGGACGCCGAGGAAGCCGGCGCCGACGGTATCACCGTGCACCTGCGCGAGGACCGCCGGCATATCCAGGAACGCGACGTGCGGGTGCTCAAGGAAGTGCTGCAGACGCGGATGAACTTCGAGATGGGGGTCACCGAGGAAATGCTCGCCTTCGCCGAGGAGATCCGGCCGGCGCATAGCTGCCTGGTCCCCGAGCGCCGCGAGGAACTGACCACCGAGGGCGGCCTCGACGTCGCCGGCCAGGAACAGCGCATCCGCGACGCGGTGCGACGCCTGGCTGCGGTGGGCAGCGAAGTGTCGCTGTTCATCGATCCCGATCCGCGGCAGATCGAGGCCTCCGCCCGGGTCGGCGCGCCGGCTATCGAACTGCATACCGGTCGCTACGCCGACGCCGAAGACCCCGAGGAGCAGGCCCGCGAGCTGCAGCGGGTACGCGAGGGCGTGGCCCTCGGGCGTTCCCTCGGCCTGATCGTCAACGCCGGGCATGGCCTGCACTACCACAATGTCGAGCCGGTCGCGGCCATCGACGGCATCAACGAGCTGAACATCGGCCACGCCATCGTCGCCCACGCCCTCTTCGTCGGCTTCAGGCAGGCGGTGGCCGAGATGAAAGCGCTGATGCTGGCCGCGGCGACCAAGCGCTGA |
| Protein Properties |
| Protein Residues: |
248 |
| Protein Molecular Weight: |
27.2 kDa |
| Protein Theoretical pI: |
5.59 |
| Hydropathicity (GRAVY score): |
-0.237 |
| Charge at pH 7 (predicted): |
-7.61 |
| Protein Sequence: |
>PA0773
MTEATRILLGVNIDHVATLRQARGTRYPDPVKAALDAEEAGADGITVHLREDRRHIQERDVRVLKEVLQTRMNFEMGVTEEMLAFAEEIRPAHSCLVPERREELTTEGGLDVAGQEQRIRDAVRRLAAVGSEVSLFIDPDPRQIEASARVGAPAIELHTGRYADAEDPEEQARELQRVREGVALGRSLGLIVNAGHGLHYHNVEPVAAIDGINELNIGHAIVAHALFVGFRQAVAEMKALMLAAATKR |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA0773 and its homologs
|