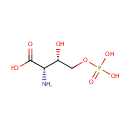| Identification |
| Name: |
4-hydroxythreonine-4-phosphate dehydrogenase |
| Synonyms: |
- 4-(phosphohydroxy)-L-threonine dehydrogenase
|
| Gene Name: |
pdxA |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in 4-hydroxythreonine-4-phosphate dehydrogenase activity |
| Specific Function: |
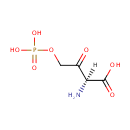
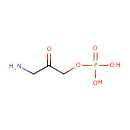
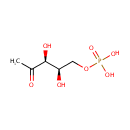
Catalyzes the NAD(P)-dependent oxidation of 4- (phosphohydroxy)-L-threonine (HTP) into 2-amino-3-oxo-4- (phosphohydroxy)butyric acid which spontaneously decarboxylates to form 3-amino-2-oxopropyl phosphate (AHAP) |
| Cellular Location: |
Cytoplasm |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| 4-hydroxythreonine-4-phosphate dehydrogenase activity | | binding | | catalytic activity | | NAD or NADH binding | | nucleotide binding | | oxidoreductase activity | | oxidoreductase activity, acting on CH-OH group of donors | | oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor | | Process |
|---|
| metabolic process | | oxidation reduction | | pyridoxine biosynthetic process | | pyridoxine metabolic process | | small molecule metabolic process | | vitamin B6 metabolic process | | vitamin metabolic process | | water-soluble vitamin metabolic process |
|
| Gene Properties |
| Locus tag: |
PA0593 |
| Strand: |
- |
| Entrez Gene ID: |
880733 |
| Accession: |
NP_249284.1 |
| GI: |
15595790 |
| Sequence start: |
651497 |
| Sequence End: |
652483 |
| Sequence Length: |
986 |
| Gene Sequence: |
>PA0593
GTGAGCCTGCGCTTCGCGCTCACTCCCGGCGAGCCGGCAGGTATCGGACCCGATCTGTGCCTGCTGCTCGCCCGCTCCGCCCAGCCCCACCCCCTGATCGCCATCGCCAGCCGCACGCTCCTGCAAGAGCGCGCCGGCCAACTGGGCCTGGCGATCGACCTTAAGGACGTTTCCCCGGCCGCCTGGCCAGAGCGACCGGCCAAGGCCGGCCAACTGTACGTATGGGATACGCCCCTGGCCGCCCCGGTCCGGCCCGGGCAATTGGACCGAGCCAACGCCGCCTACGTCCTGGAAACCCTGACCCGCGCCGGCCAGGGCTGCCTCGACGGGCACTTCGCCGGGATGATCACCGCACCGGTGCACAAGGGCGTGATCAACGAAGCCGGCATCCCCTTTTCCGGCCATACCGAATTCCTCGCCGACCTCACCCACACCGCCCAGGTGGTGATGATGCTGGCCACCCGCGGCCTGCGCGTGGCCCTGGCGACCACCCACCTGCCGCTGCGCGAGGTCGCCGACGCCATTAGCGATGAGCGCCTGACCCGGGTCGCGCGCATCCTCCACGCCGACCTGCGCGACAAGTTCGGCATCGCCCATCCGCGCATCCTGGTCTGCGGACTCAACCCGCACGCCGGCGAAGGCGGTCACCTGGGCCGGGAAGAGATCGAGGTGATCGAACCCTGCCTCGAGCGCCTGCGTGGCGAAGGCCTCGACCTGATCGGCCCGCTGCCGGCCGACACCCTGTTCACTCCCAAGCACCTGGAGCATTGCGACGCGGTGCTGGCGATGTACCACGACCAGGGTCTCCCCGTGCTCAAGTACAAGGGCTTCGGTGCCGCCGTGAACGTGACCCTCGGCCTGCCGATCATCCGCACCTCTGTCGACCACGGTACCGCGCTCGACCTGGCCGGCAGCGGCCGCATCGATAGCGGCAGCCTGCAGGTCGCCCTGGAAACCGCCTACCAGATGGCGGCCAGCCGCTGCTGA |
| Protein Properties |
| Protein Residues: |
328 |
| Protein Molecular Weight: |
34.9 kDa |
| Protein Theoretical pI: |
6.62 |
| Hydropathicity (GRAVY score): |
0.121 |
| Charge at pH 7 (predicted): |
-2.81 |
| Protein Sequence: |
>PA0593
MSLRFALTPGEPAGIGPDLCLLLARSAQPHPLIAIASRTLLQERAGQLGLAIDLKDVSPAAWPERPAKAGQLYVWDTPLAAPVRPGQLDRANAAYVLETLTRAGQGCLDGHFAGMITAPVHKGVINEAGIPFSGHTEFLADLTHTAQVVMMLATRGLRVALATTHLPLREVADAISDERLTRVARILHADLRDKFGIAHPRILVCGLNPHAGEGGHLGREEIEVIEPCLERLRGEGLDLIGPLPADTLFTPKHLEHCDAVLAMYHDQGLPVLKYKGFGAAVNVTLGLPIIRTSVDHGTALDLAGSGRIDSGSLQVALETAYQMAASRC |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA0593 and its homologs
|