|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB002046 |
|---|
|
Identification |
|---|
| Name: |
Ferricytochrome c |
|---|
| Description: | Cytochrome c, or Cyt c, is a small heme protein and a component of the oxidative phosphorylation electron transport chain. The heme group of cytochrome c accepts electrons from the Cytochrome b-c1 complex (Complex III) and transfers electrons to the Cytochrome oxidase complex (Complex IV). Cyt c is capable of undergoing oxidation and reduction, but does not bind oxygen. Cytochrome c is a highly conserved protein across the spectrum of species, found in plants, animals, and many unicellular organisms. This, along with its small size (molecular weight about 12,000 daltons), makes it useful in studies of cladistics. Its primary structure consists of a chain of about 100 amino acids. (Wikipedia) |
|---|
|
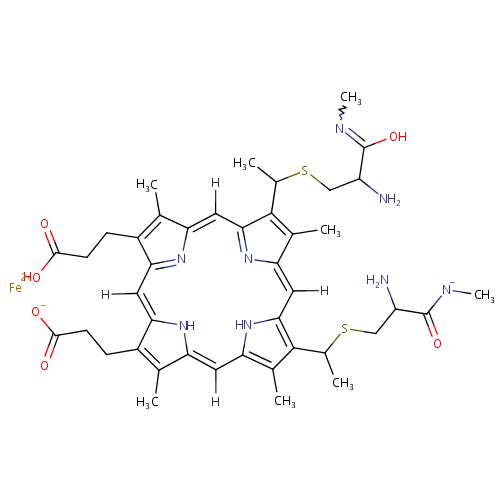
Structure |
|
|---|
| Synonyms: | - Cytochrome c
- Cytochrome c (JAN)
- Ferricytochrome c
- Holocytochrome c
|
|---|
|
Chemical Formula: |
C42H68O13 |
|---|
| Average Molecular Weight: |
780.993 |
|---|
| Monoisotopic Molecular
Weight: |
780.46599225 |
|---|
| InChI Key: |
FHOKVOIILRHONR-ZVBGSRNCSA-N |
|---|
| InChI: | InChI=1S/C42H68O13/c1-21(19-52-37-36(51)34(49)32(47)27(55-37)20-53-38-35(50)33(48)31(46)26(18-43)54-38)9-8-10-22(2)23-15-16-40(5)28-13-11-24-25(12-14-29(44)39(24,3)4)42(28,7)30(45)17-41(23,40)6/h9,11,22-23,25-29,31-38,43-44,46-51H,8,10,12-20H2,1-7H3/b21-9+ |
|---|
| CAS
number: |
9007-43-6 |
|---|
| IUPAC Name: | ??-iron(2+) ion 3-[15-(1-{[2-amino-2-(methyl-C-hydroxycarbonimidoyl)ethyl]sulfanyl}ethyl)-10-(1-{[2-amino-3-(methylazanidyl)-3-oxopropyl]sulfanyl}ethyl)-20-(2-carboxyethyl)-5,9,14,19-tetramethyl-21,22,23,24-tetraazapentacyclo[16.2.1.1?,??1????.1??,???tetracosa-1(21),2,4,6,8,10,12,14,16(22),17,19-undecaen-4-yl]propanoate |
|---|
|
Traditional IUPAC Name: |
??-iron(2+) ion 3-[15-(1-{[2-amino-2-(methyl-C-hydroxycarbonimidoyl)ethyl]sulfanyl}ethyl)-10-(1-{[2-amino-3-(methylazanidyl)-3-oxopropyl]sulfanyl}ethyl)-20-(2-carboxyethyl)-5,9,14,19-tetramethyl-21,22,23,24-tetraazapentacyclo[16.2.1.1?,??1????.1??,???tetracosa-1(21),2,4,6,8,10,12,14,16(22),17,19-undecaen-4-yl]propanoate |
|---|
| SMILES: | CC(CC\C=C(/C)COC1OC(COC2OC(CO)C(O)C(O)C2O)C(O)C(O)C1O)C1CCC2(C)C3CC=C4C(CCC(O)C4(C)C)C3(C)C(=O)CC12C |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as metalloporphyrins. These are polycyclic compounds containing a porphyrin moiety and a metal atom. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
|
Class |
Tetrapyrroles and derivatives |
|---|
| Sub Class | Metallotetrapyrroles |
|---|
|
Direct Parent |
Metalloporphyrins |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Metalloporphyrin
- Porphyrin
- Alpha-amino acid amide
- Alpha-amino acid or derivatives
- Substituted pyrrole
- Dicarboxylic acid or derivatives
- Heteroaromatic compound
- Pyrrole
- Carboxylic acid salt
- Carboxamide group
- Azacycle
- Dialkylthioether
- Organic 1,3-dipolar compound
- Carbene-type 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Sulfenyl compound
- Thioether
- Carboxylic acid
- Carboxylic acid derivative
- Carboximidic acid derivative
- Carboximidic acid
- Hydrocarbon derivative
- Organic transition metal salt
- Organic salt
- Primary amine
- Organosulfur compound
- Organooxygen compound
- Organonitrogen compound
- Primary aliphatic amine
- Carbonyl group
- Amine
- Organic zwitterion
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework |
Aromatic heteropolycyclic compounds |
|---|
| External Descriptors |
Not Available |
|---|
|
Physical Properties |
|---|
| State: |
Solid |
|---|
| Charge: | -1 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Cytoplasm |
|---|
| Reactions: | |
|---|
|
Pathways: |
|
|---|
|
Spectra |
|---|
| Spectra: |
|
|---|
|
References |
|---|
| References: |
Not Available |
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|