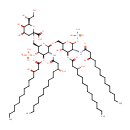| Identification |
| Name: |
3-deoxy-D-manno-octulosonic-acid transferase |
| Synonyms: |
|
| Gene Name: |
waaA |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in biosynthetic process |
| Specific Function: |
Essential step in lipopolysaccharides biosynthesis. Acts at transfer of 3-deoxy-D-mono octulonic acid (KDO) from CMP-KDO to a tetraacyldisaccharide 1,4'-bisphosphate precursor of lipid A (lipid IVA). Transfers two molecules of KDO to lipid IVA. Degraded by FtsH; therefore FtsH regulates the addition of the sugar moiety of the LPS and thus the maturation of the LPS precursor |
| Cellular Location: |
Cell inner membrane; Single-pass membrane protein (Probable) |
| KEGG Pathways: |
|
| KEGG Reactions: |
| | |
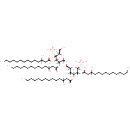
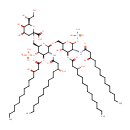 | + | 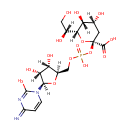 | ↔ | Di[3-deoxy-D-manno-octulosonyl]-lipid IV(A) | + | 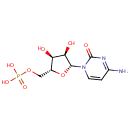 |
| | |
Di[3-deoxy-D-manno-octulosonyl]-lipid IV(A) | + |  | ↔ | alpha-Kdo-(2->8)-alpha-Kdo-(2->4)-alpha-Kdo-(2->6)-lipid IVA | + |  |
| |
|
| SMPDB Reactions: |
|
 | + |  | → | Cytidine monophosphate | + |  | + |  | + |  |
| | |
 | + |  | → | Cytidine monophosphate | + |  | + | a-Kdo-(2->4)-a-Kdo-(2->6)-lipid IVA | + |  | + |  |
| |
|
| PseudoCyc/BioCyc Reactions: |
| | |
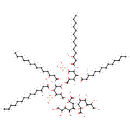
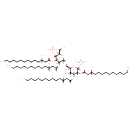
 | + |  | ↔ |  | + | α-Kdo-(2->4)-α-Kdo-(2->6)-lipid IVA | + |  |
| |
|
| Complex Reactions: |
| | | | |
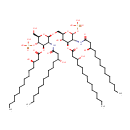 | + |  | → | alpha-Kdo-(2->6)-lipid IV(A) | + |  |
| | |
Alpha-Kdo-(2->6)-lipid IV(A) | + |  | → | alpha-Kdo-(2->4)-alpha-Kdo-(2->6)-lipid IV(A) | + |  |
| |
|
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
| PAMDB003481 | (2-N,3-O-bis(3-Hydroxytetradecanoyl)-4-O-phosphono-beta-D-glucosaminyl)-(1->6)-(2-N,3-O-bis(3-hydroxytetradecanoyl)-beta-D-glucosaminyl phosphate) | MetaboCard | | PAMDB000906 | 2,3,2'3'-Tetrakis(3-hydroxytetradecanoyl)-D-glucosaminyl-1,6-beta-D-glucosamine 1,4'-bisphosphate | MetaboCard | | PAMDB001545 | 2,3,2'3'-Tetrakis(beta-hydroxymyristoyl)-D-glucosaminyl-1,6-beta-D-glucosamine 1,4'-bisphosphate | MetaboCard | | PAMDB006295 | a-Kdo-(2->4)-a-Kdo-(2->6)-lipid IVA | MetaboCard | | PAMDB006335 | alpha-Kdo-(2??)-lipid IVA | MetaboCard | | PAMDB000981 | CMP-3-Deoxy-D-manno-octulosonate | MetaboCard | | PAMDB000035 | Cytidine monophosphate | MetaboCard | | PAMDB001633 | Hydrogen ion | MetaboCard | | PAMDB001643 | KDO(2)-lipid IV(A) | MetaboCard | | PAMDB001644 | KDO-lipid IV(A) | MetaboCard |
|
| GO Classification: |
| Function |
|---|
| binding | | carbohydrate binding | | catalytic activity | | sugar binding | | transferase activity | | Process |
|---|
| biosynthetic process | | carbohydrate metabolic process | | metabolic process | | primary metabolic process |
|
| Gene Properties |
| Locus tag: |
PA4988 |
| Strand: |
- |
| Entrez Gene ID: |
880145 |
| Accession: |
NP_253675.1 |
| GI: |
15600181 |
| Sequence start: |
5603821 |
| Sequence End: |
5605098 |
| Sequence Length: |
1277 |
| Gene Sequence: |
>PA4988
ATGAACCGTACCCTCTATACCCTGCTGTTCCACCTCGGCTTGCCGCTGGTGGCGTTGCGCCTGTGGTGGCGCGCGCGCCAGGCGCCGGCCTATGCCAAGCGCATCGGCGAGCGTTTTTCCCTGTCCTTGCCCGAGGTGCCGCCCGGCGGCATCTGGGTGCACGCCGTCTCGGTGGGCGAGAGCATCGCCGCCGCGCCGATGGTGCGGGCGCTGCTGGAGCGCCATCCGCAGCTGCCGGTCACCGTCACCTGCATGACCCCGACCGGCTCGGAGCGCATCCGCGCATTGTTCGGCGAGCAGGTCCGGCACTGCTACCTGCCCTACGACCTGCCCTGGGCGGCGGCGCGCTTCCTCGACCGGGTGCGGCCGCGCCTGGCGGTGATCATGGAAACCGAGCTGTGGCCCAACCATATCCATGCCTGCGCCGTGCGCGGGATCCCGGTGGCGCTGGCCAATGCGCGCCTGTCCGAGCGCTCGGCGCGTGGCTACGCGCGCTTCGCCGGGCTGACCCGGCCGATGCTCGCCGAGCTGTCCTGGATCGCCGTGCAGACCGAGGCCGAGGCCGAGCGCTTCCGTCGTCTCGGCGCGCGTCCTGAGTGCGTCAGCGTGACCGGCTCGATCAAGTTCGACCTGCGCATCGACCCGCAGCTGCCGCTGGCCGCTGCCGCCCTGCGCGAGGAATGGGACGCGACGGCGCGACCGCTGTGGATCGCCGCCAGCACCCATGCCGGCGAGGACGAGATCGTCCTGGCAGCCCACCGGCGCCTGCTGGAAACGCGTCCCGACGCGTTGCTGATCCTGGTGCCGCGCCATCCGGAACGCTTCGCCGGCGTCCACGAGCTATGCCGCCGCGAAGGCTTCGCCACGGTCCGGCGCTCCGGCGGCGAGCCGGTGGCACGCGCGACCCAGGTGCTGCTCGGCGATACCATGGGCGAGCTGCTGTTCCTCTATGCCCTGGCCGACATCGCTTTCGTCGGCGGCAGCCTGGTGCCCAACGGCGGGCACAACCTGCTGGAGCCGGCGGCCCTGGGCAAGCCGGTGTTCGCCGGTCCGCACCTGTTCAACTTCCTCGACATCGCCGCGCAATTGCGCGACGCCGGGGCGCTGCTCGAGGTGACGGATGCCGGCGAGCTGTGCGACGGCCTCGCGCGCCTGTGGGCGCAGCCGGAGGTCGCGACGGCCATGGCCACTGCCGGAGAGAAGGTGCTGCGCAACAACCAGGGCGCCCTGGAGCGGCTGCTGGCGGGGTTGGCGCGGTTGCTCGGGCGCGGCGGGTAG |
| Protein Properties |
| Protein Residues: |
425 |
| Protein Molecular Weight: |
46.5 kDa |
| Protein Theoretical pI: |
8.83 |
| Hydropathicity (GRAVY score): |
0.102 |
| Charge at pH 7 (predicted): |
7.72 |
| Protein Sequence: |
>PA4988
MNRTLYTLLFHLGLPLVALRLWWRARQAPAYAKRIGERFSLSLPEVPPGGIWVHAVSVGESIAAAPMVRALLERHPQLPVTVTCMTPTGSERIRALFGEQVRHCYLPYDLPWAAARFLDRVRPRLAVIMETELWPNHIHACAVRGIPVALANARLSERSARGYARFAGLTRPMLAELSWIAVQTEAEAERFRRLGARPECVSVTGSIKFDLRIDPQLPLAAAALREEWDATARPLWIAASTHAGEDEIVLAAHRRLLETRPDALLILVPRHPERFAGVHELCRREGFATVRRSGGEPVARATQVLLGDTMGELLFLYALADIAFVGGSLVPNGGHNLLEPAALGKPVFAGPHLFNFLDIAAQLRDAGALLEVTDAGELCDGLARLWAQPEVATAMATAGEKVLRNNQGALERLLAGLARLLGRGG |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA4988 and its homologs
|