| Identification |
| Name: |
NH(3)-dependent NAD(+) synthetase |
| Synonyms: |
- Nicotinamide adenine dinucleotide synthetase
- NADS
- Nitrogen regulatory protein
|
| Gene Name: |
nadE |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in NAD+ synthase (glutamine-hydrolyzing) activity |
| Specific Function: |
Catalyzes a key step in NAD biosynthesis, transforming deamido-NAD into NAD by a two-step reaction |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
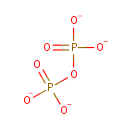
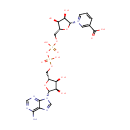 | + |  | + | 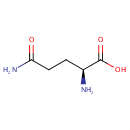 | + | 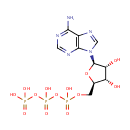 | → |  | + | 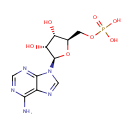 | + | Pyrophosphate | + | L-Glutamic acid | + | 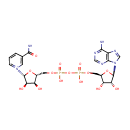 | + | 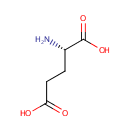 |
| | | |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| adenyl nucleotide binding | | adenyl ribonucleotide binding | | ATP binding | | binding | | carbon-nitrogen ligase activity, with glutamine as amido-N-donor | | catalytic activity | | ligase activity | | ligase activity, forming carbon-nitrogen bonds | | NAD+ synthase (glutamine-hydrolyzing) activity | | nucleoside binding | | purine nucleoside binding | | Process |
|---|
| cellular metabolic process | | coenzyme biosynthetic process | | coenzyme metabolic process | | cofactor metabolic process | | metabolic process | | NAD biosynthetic process | | nicotinamide nucleotide biosynthetic process | | pyridine nucleotide biosynthetic process |
|
| Gene Properties |
| Locus tag: |
PA4920 |
| Strand: |
+ |
| Entrez Gene ID: |
882213 |
| Accession: |
NP_253607.1 |
| GI: |
15600113 |
| Sequence start: |
5519711 |
| Sequence End: |
5520538 |
| Sequence Length: |
827 |
| Gene Sequence: |
>PA4920
ATGCAACAGATCCAACGCGACATCGCCCAAGCCCTGCAGGTCCAGCCGCCGTTCCAGTCGGAGGCCGACGTGCAGGCGCAGATCGCCCGGCGCATCGCCTTCATCCAGCAGTGCCTGAAGGATTCCGGGCTGAAGACCCTGGTGCTGGGGATCAGCGGCGGCGTCGACTCGCTCACCGCCGGCCTGCTGGCCCAGCGCGCCGTCGAGCAACTGCGCGAGCAGACCGGCGACCAGGCCTACCGCTTCATCGCCGTGCGCCTGCCCTACCAGGTGCAGCAGGACGAGGCCGATGCCCAGGCCTCGCTGGCGACCATCCGCGCCGACGAAGAGCAGACCGTCAACATCGGTCCGTCGGTGAAAGCCCTGGCGGAACAGCTGGAAGCCCTGGAAGGACTCGAGCCGGCGAAGAGCGACTTCGTCATCGGCAATATCAAGGCGCGCATCCGCATGGTCGCCCAGTACGCCATCGCCGGCGCCCGCGGCGGCCTGGTGATCGGCACCGACCATGCTGCCGAGGCGGTCATGGGGTTCTTCACCAAGTTCGGCGACGGCGCCTGCGACCTGGCTCCGCTCAGCGGCCTGGCCAAGCATCAGGTACGCGCCCTCGCCCGCGCCCTCGGCGCTCCGGAGAACCTGGTGGAGAAGATCCCCACCGCCGACCTCGAGGACCTGCGCCCCGGCCATCCGGACGAGGCTTCCCACGGCGTCACCTATGCCGAGATCGACGCCTTCCTGCACGGCCAGCCGCTGCGCGAGGAAGCTGCGCGGGTGATCGTCGACACCTACCACAAGACCCAGCACAAGCGCGAACTGCCGAAGGCGCCCTGA |
| Protein Properties |
| Protein Residues: |
275 |
| Protein Molecular Weight: |
29.7 kDa |
| Protein Theoretical pI: |
5.38 |
| Hydropathicity (GRAVY score): |
-0.193 |
| Charge at pH 7 (predicted): |
-6.37 |
| Protein Sequence: |
>PA4920
MQQIQRDIAQALQVQPPFQSEADVQAQIARRIAFIQQCLKDSGLKTLVLGISGGVDSLTAGLLAQRAVEQLREQTGDQAYRFIAVRLPYQVQQDEADAQASLATIRADEEQTVNIGPSVKALAEQLEALEGLEPAKSDFVIGNIKARIRMVAQYAIAGARGGLVIGTDHAAEAVMGFFTKFGDGACDLAPLSGLAKHQVRALARALGAPENLVEKIPTADLEDLRPGHPDEASHGVTYAEIDAFLHGQPLREEAARVIVDTYHKTQHKRELPKAP |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA4920 and its homologs
|