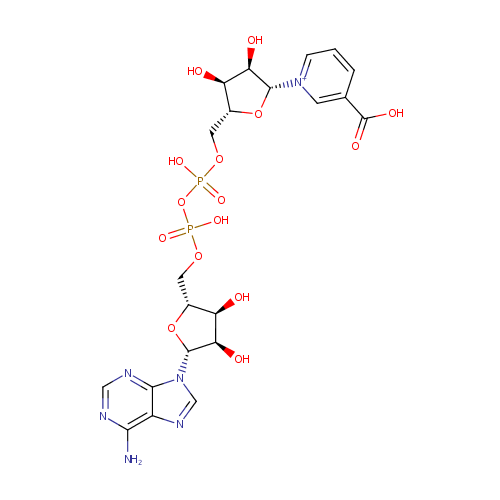
Nicotinic acid adenine dinucleotide (PAMDB000286)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000286 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Nicotinic acid adenine dinucleotide | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Nicotinic acid adenine dinucleotide phosphate, (NAADP), is a Ca2+-mobilizing second messenger synthesized in response to extracellular stimuli. NAADP binds to and opens Ca2+ channels on intracellular organelles, thereby increasing the intracellular Ca2+ concentration which, in turn, modulates a variety of cellular processes. Structurally, it is a dinucleotide that only differs from the house-keeping enzyme cofactor, NADP by a hydroxyl group (replacing the nicotinamide amino group) and yet this minor modification converts it into the most potent Ca2+-mobilizing second messenger yet described. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C21H27N6O15P2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 665.4178 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 665.100962248 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | SENPVEZBRZQVST-HISDBWNOSA-O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C21H26N6O15P2/c22-17-12-18(24-7-23-17)27(8-25-12)20-16(31)14(29)11(41-20)6-39-44(36,37)42-43(34,35)38-5-10-13(28)15(30)19(40-10)26-3-1-2-9(4-26)21(32)33/h1-4,7-8,10-11,13-16,19-20,28-31H,5-6H2,(H4-,22,23,24,32,33,34,35,36,37)/p+1/t10-,11-,13-,14-,15-,16-,19-,20-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 6450-77-7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 1-[(2R,3R,4S,5R)-5-[({[({[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl)oxy](hydroxy)phosphoryl}oxy)methyl]-3,4-dihydroxyoxolan-2-yl]-3-carboxy-1???pyridin-1-ylium | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | deamido-NAD(+) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | NC1=C2N=CN([C@@H]3O[C@H](COP(O)(=O)OP(O)(=O)OC[C@H]4O[C@H]([C@H](O)[C@@H]4O)[N+]4=CC=CC(=C4)C(O)=O)[C@@H](O)[C@H]3O)C2=NC=N1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as purine nucleotide sugars. These are purine nucleotides bound to a saccharide derivative through the terminal phosphate group. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Nucleosides, nucleotides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Purine nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Purine nucleotide sugars | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Purine nucleotide sugars | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Hydrogen ion + Nicotinamide ribotide <> Nicotinic acid adenine dinucleotide + Pyrophosphate Adenosine triphosphate + Nicotinic acid adenine dinucleotide + Ammonium > Adenosine monophosphate + Hydrogen ion + NAD + Pyrophosphate Adenosine triphosphate + Nicotinic acid adenine dinucleotide + Ammonia <> Adenosine monophosphate + Pyrophosphate + NAD Nicotinic acid adenine dinucleotide + Water <> Adenosine monophosphate + Nicotinamide ribotide Adenosine triphosphate + Nicotinamide ribotide <> Pyrophosphate + Nicotinic acid adenine dinucleotide Adenosine triphosphate + Nicotinic acid adenine dinucleotide + L-Glutamine + Water > Hydrogen ion + Adenosine monophosphate + Pyrophosphate + NAD + L-Glutamate nicotinate beta-D-ribonucleotide + Adenosine triphosphate + Hydrogen ion + Nicotinamide ribotide > Pyrophosphate + Nicotinic acid adenine dinucleotide Nicotinic acid adenine dinucleotide + Water + L-Glutamine + Adenosine triphosphate > Hydrogen ion + Adenosine monophosphate + Pyrophosphate + L-Glutamic acid + NAD + L-Glutamate Nicotinic acid adenine dinucleotide + Adenosine triphosphate + Ammonium > Hydrogen ion + Adenosine monophosphate + Pyrophosphate + NAD | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||