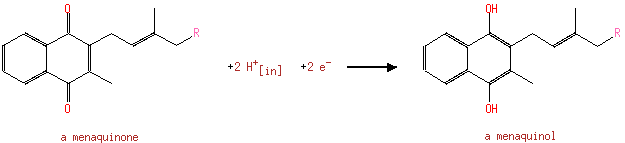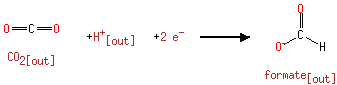| Identification |
| Name: |
Formate dehydrogenase, nitrate-inducible, iron-sulfur subunit |
| Synonyms: |
- Anaerobic formate dehydrogenase iron-sulfur subunit
- Formate dehydrogenase-N subunit beta
- FDH-N subunit beta
|
| Gene Name: |
fdnH |
| Enzyme Class: |
Not Available |
| Biological Properties |
| General Function: |
Involved in electron carrier activity |
| Specific Function: |
Formate dehydrogenase allows E.coli to use formate as major electron donor during anaerobic respiration, when nitrate is used as electron acceptor. The beta chain is an electron transfer unit containing 4 cysteine clusters involved in the formation of iron-sulfur centers. Electrons are transferred from the gamma chain to the molybdenum cofactor of the alpha subunit |
| Cellular Location: |
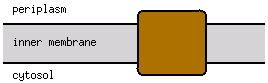
Cell membrane; Single-pass membrane protein |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
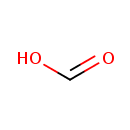
| | | formate [inner membrane] + NAD +[inner membrane] → CO 2[inner membrane] + NADH [inner membrane] + H +[inner membrane]ReactionCard | | a menaquinone [membrane] + 2 H +[in] + 2 e -[membrane] → a menaquinol [membrane]ReactionCard | | CO 2[out] + H +[out] + 2 e -[membrane] → formate [out]ReactionCard | |
 | + | ETR-Quinones | + |  | → | 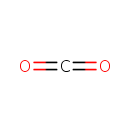 | + | ETR-Quinols |
| |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Component |
|---|
| cell part | | integral to membrane | | intrinsic to membrane | | membrane part | | Function |
|---|
| binding | | catalytic activity | | electron carrier activity | | formate dehydrogenase activity | | iron-sulfur cluster binding | | metal cluster binding | | oxidoreductase activity | | oxidoreductase activity, acting on the aldehyde or oxo group of donors | | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor | | Process |
|---|
| cellular metabolic process | | cellular respiration | | energy derivation by oxidation of organic compounds | | generation of precursor metabolites and energy | | metabolic process |
|
| Gene Properties |
| Locus tag: |
PA4811 |
| Strand: |
- |
| Entrez Gene ID: |
880193 |
| Accession: |
NP_253499.1 |
| GI: |
15600005 |
| Sequence start: |
5397690 |
| Sequence End: |
5398619 |
| Sequence Length: |
929 |
| Gene Sequence: |
>PA4811
ATGAGTAGTCAGGACATCCACGCCCGCTCCGCGACCACCACGCCGATGCCCTCGGTGCGGAACCAGGAGGAAGTGGCGAAACTGATCGACGTGTCCAAGTGCATCGGTTGCAAGGCCTGCCAGGTCGCCTGCTCGGAATGGAACGAGCTGCGCGACGAGGTCGGCCACAACCACGGCACCTACGACAACCCCATGGACCTGACGGCCGACTCGTGGACGGTGATGCGTTTCACCGAGCACGAGAACGAGGCCGGCAACCTCGAGTGGCTGATCCGCAAGGATGGCTGCATGCACTGCGCCGAGCCGGGCTGCCTGAAGGCCTGCCCGAGCCCGGGAGCGATCGTGAAGTACGCCAACGGCATCGTCGACTTCAACCAGGACAAGTGCATCGGTTGCGGCTACTGCATCACCGGTTGCCCGTTCGACATCCCACGGATCTCGCAGAAGGACCACAAGGCCTACAAGTGCAGCCTGTGTTCCGACCGCGTCTCGGTGGGCATGGAGCCGGCCTGCGTGAAGACCTGCCCGACCGGCGCCATCGTCTTCGGCAGCAAGGAGGACATGAAGGAACACGCCGCCGAACGCATCGTCGACCTGAAGAGCCGCGGTTACGAGAACGCCGGCCTGTACGACCCCGAAGGGGTGGGCGGCACCCACGTGATGTACGTGCTGCACCACGCCGACAAGCCGTCGCTCTATGCCGGCCTGGCCGACCAGCCGAGCGTCAGCCCGCTGGTCAGCCTGTGGAAGGGCGTGACCAAGCCGCTGGCGCTGCTGGCCATGGGCGCCACGGTGCTCGCCGGGTTCTTCCACTATGTGCGCGTCGGTCCCAACCGCGCGGACGACGAGGAAGACCACGGGCAAGCCGAGGCACCGGTGCACCAGGTCGATCCGTCGGTACACACCTTTGACCCGCACGAGCGTCGTTGA |
| Protein Properties |
| Protein Residues: |
309 |
| Protein Molecular Weight: |
33.8 kDa |
| Protein Theoretical pI: |
5.97 |
| Hydropathicity (GRAVY score): |
-0.359 |
| Charge at pH 7 (predicted): |
-8.88 |
| Protein Sequence: |
>PA4811
MSSQDIHARSATTTPMPSVRNQEEVAKLIDVSKCIGCKACQVACSEWNELRDEVGHNHGTYDNPMDLTADSWTVMRFTEHENEAGNLEWLIRKDGCMHCAEPGCLKACPSPGAIVKYANGIVDFNQDKCIGCGYCITGCPFDIPRISQKDHKAYKCSLCSDRVSVGMEPACVKTCPTGAIVFGSKEDMKEHAAERIVDLKSRGYENAGLYDPEGVGGTHVMYVLHHADKPSLYAGLADQPSVSPLVSLWKGVTKPLALLAMGATVLAGFFHYVRVGPNRADDEEDHGQAEAPVHQVDPSVHTFDPHERR |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA4811 and its homologs
|