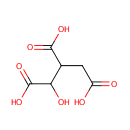
Isocitric acid (PAMDB000604)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000604 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Isocitric acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Isocitric acid is a protonated form of isocitrate, which is a substrate of the citric acid cycle. Isocitrate is formed from citrate with the help of the enzyme aconitase, and is acted upon by isocitrate dehydrogenase. Salts and esters of isocitric acid are known as isocitrates. (Wikipedia) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C6H8O7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 192.1235 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 192.02700261 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | ODBLHEXUDAPZAU-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C6H8O7/c7-3(8)1-2(5(10)11)4(9)6(12)13/h2,4,9H,1H2,(H,7,8)(H,10,11)(H,12,13) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 320-77-4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 1-hydroxypropane-1,2,3-tricarboxylic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | isocitric acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OC(C(CC(O)=O)C(O)=O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as tricarboxylic acids and derivatives. These are carboxylic acids containing exactly three carboxyl groups. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Tricarboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Tricarboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 162-165 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | cis-Aconitic acid + Water <> Isocitric acid Isocitric acid + NADP <> alpha-Ketoglutarate + Carbon dioxide + NADPH Isocitric acid <> Glyoxylic acid + Succinic acid Isocitric acid + NADP <> alpha-Ketoglutarate + Carbon dioxide + NADPH + Hydrogen ion Citric acid <> Isocitric acid Isocitric acid + NADP <> Oxalosuccinic acid + NADPH + Hydrogen ion Isocitric acid + NADP > NADPH + Oxoglutaric acid + Carbon dioxide Citric acid + cis-Aconitic acid + Water <> Isocitric acid Isocitric acid + NADP + Oxalosuccinic acid <> alpha-Ketoglutarate + Carbon dioxide + NADPH + Hydrogen ion Isocitric acid + NAD + Isocitric acid > Oxoglutaric acid + Carbon dioxide + NADH + Hydrogen ion cis-Aconitic acid + Water <> Isocitric acid + Isocitric acid Isocitric acid + Isocitric acid <> Succinic acid + Glyoxylic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Finogenova, T. V.; Kamzolova, S. V.; Dedyukhina, E. G.; Shishkanova, N. V.; Il'chenko, A. P.; Morgunov, I. G.; Chernyavskaya, O. G.; Sokolov, A. P. Biosynthesis of citric and isocitric acids from ethanol by mutant Yarrowia lipolytica N 1 under continuous | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in magnesium ion binding
- Specific function:
- Isocitrate + NADP(+) = 2-oxoglutarate + CO(2) + NADPH
- Gene Name:
- icd
- Locus Tag:
- PA2623
- Molecular weight:
- 45.6 kDa
Reactions
| Isocitrate + NADP(+) = 2-oxoglutarate + CO(2) + NADPH. |
- General function:
- Involved in isocitrate lyase activity
- Specific function:
- Catalyzes the formation of succinate and glyoxylate from isocitrate, a key step of the glyoxylate cycle. May be involved in the assimilation of one-carbon compounds via the isocitrate lyase- positive serine pathway
- Gene Name:
- aceA
- Locus Tag:
- PA2634
- Molecular weight:
- 58.9 kDa
Reactions
| Isocitrate = succinate + glyoxylate. |
- General function:
- Involved in metabolic process
- Specific function:
- May have an iron-responsive regulatory function
- Gene Name:
- acnA
- Locus Tag:
- PA1562
- Molecular weight:
- 99.1 kDa
Reactions
| Citrate = isocitrate. |
- General function:
- Involved in metabolic process
- Specific function:
- Citrate = isocitrate
- Gene Name:
- acnB
- Locus Tag:
- PA1787
- Molecular weight:
- 93.6 kDa
Reactions
| Citrate = isocitrate. |
| (2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate = (Z)-but-2-ene-1,2,3-tricarboxylate + H(2)O. |