| Identification |
| Name: |
Glutamate synthase [NADPH] small chain |
| Synonyms: |
- Glutamate synthase subunit beta
- GLTS beta chain
- NADPH-GOGAT
|
| Gene Name: |
gltD |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in iron-sulfur cluster binding |
| Specific Function: |
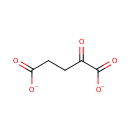
2 L-glutamate + NADP(+) = L-glutamine + 2- oxoglutarate + NADPH |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
- Alanine, aspartate and glutamate metabolism pae00250
- Metabolic pathways pae01100
- Microbial metabolism in diverse environments pae01120
- Nitrogen metabolism pae00910
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
2 L-Glutamic acid | + |  | + | 2 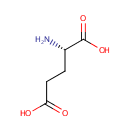 | → | 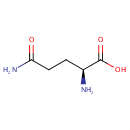 | + | NADPH | + |  | + | 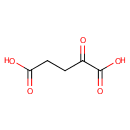 | + | 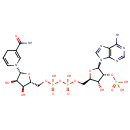 |
| | |
 | + |  | + | NADPH | + |  | + |  | → | 2 L-Glutamic acid | + |  | + | 2  |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
Not Available |
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| adenyl nucleotide binding | | binding | | catalytic activity | | FAD or FADH2 binding | | iron-sulfur cluster binding | | metal cluster binding | | nucleoside binding | | oxidoreductase activity | | oxidoreductase activity, acting on the CH-NH2 group of donors | | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor | | purine nucleoside binding | | Process |
|---|
| cellular amino acid and derivative metabolic process | | cellular amino acid metabolic process | | cellular metabolic process | | glutamate biosynthetic process | | glutamate metabolic process | | glutamine family amino acid metabolic process | | metabolic process | | oxidation reduction |
|
| Gene Properties |
| Locus tag: |
PA5035 |
| Strand: |
- |
| Entrez Gene ID: |
881221 |
| Accession: |
NP_253722.1 |
| GI: |
15600228 |
| Sequence start: |
5666234 |
| Sequence End: |
5667667 |
| Sequence Length: |
1433 |
| Gene Sequence: |
>PA5035
ATGTCTGAACGTCTGAATTCCGACCGCCTGAACAACGACTTCCAGTTCATCGAAGTCGGGCGCAAGGACCCGAAGAAGAAGCTGCTGCGCCAGCGCAAGCGCGAGTTCGTGGAGATCCACGACCTGTTCAAGCCGCAGCAGGCCGCCGACCAGGCCCACCGCTGCCTGGGCTGCGGCAACCCGTACTGCGAGTGGAAATGCCCGGTGCACAACTTCATCCCTAACTGGCTGAAGCTGGTTTCCGAAGGCAACATCCTGGCCGCCGCCGAACTGTCGCACCAGACCAACACCCTGCCGGAAGTCTGCGGCCGGGTCTGCCCGCAGGATCGCCTCTGCGAGGGCGCCTGCACCCTCAACGACGGTTTTGGCGCGGTGACCATCGGCTCGGTGGAGAAGTACATCACCGATACGGCCTTCGCCATGGGCTGGCGTCCGGACCTGTCCAAGGTCAAGCCGACCGGCAGGCGCGTCGCGGTGATCGGCGCCGGCCCGGCCGGCCTCGGCTGCGCCGACGTGCTGGTGCGCAACGGCGTGACCCCGGTGGTGTTCGACCGCAACCCGGAGATCGGCGGCCTGCTGACCTTCGGCATTCCCGAGTTCAAGCTGGAGAAGCACGTCCTCAGCCGTCGCCGCGAGGTGTTCACCGGCATGGGCATCGAGTTTCGCCTGAACACCGAGATCGGCAAGGACGTGACCATGGAGCAGTTGCTCGATGAGTACGATGCCGTGTTCATGGGGATGGGCACCTACACCTACATGAAGGGCGGTTTCCCCGGCGAGGACCTGCCGGGCGTACACGACGCCCTGGACTTCCTGATCGCCAACGTGAACCGCAACCTCGGCTTCGAGAAGTCGGCCGAGGACTTCGTCGACATGAAGGGCAAGCGCGTGGTGGTCCTGGGCGGCGGCGACACCGCGATGGACTGCAACCGCACTTCGATCCGCCAGGGCGCCAAGTCGGTGACCTGTGCCTATCGCCGTGACGAAGAAAACATGCCGGGCTCGCGCAAGGAAGTGAAGAACGCCAAGGAAGAGGGCGTGAAGTTCCTCTTCAACCGCCAGCCCATCGCCATCGTCGGCGAGGACCGGGTGGAAGGCGTGAAGGTGGTCGAGACCCGTCTCGGCGAGCCGGACGCCCGCGGCCGCCGCAGCCCCGAACCGATCCCCGGTTCCGAGGAGATCATCCCGGCGGAAGCCGTGCTGATCGCCTTCGGCTTCCGGCCGAGTCCGGCGCCGTGGTTCGAGCGCTTCGAGATCGCCACCGACAGCCAGGGCCGCGTGGTGGCTCCGGCGCAGGCGCAGTTCAAGCACCAGACCAGCAACCCGAAGATCTTCGCCGGTGGCGACATGGTCCGTGGTTCCGACCTGGTGGTGACGGCGATCTTCGAAGGACGTACCGCGGCCGAAGGCATCCTCGACTACCTTGGCGTCTGA |
| Protein Properties |
| Protein Residues: |
477 |
| Protein Molecular Weight: |
52.6 kDa |
| Protein Theoretical pI: |
6.42 |
| Hydropathicity (GRAVY score): |
-0.313 |
| Charge at pH 7 (predicted): |
-2.62 |
| Protein Sequence: |
>PA5035
MSERLNSDRLNNDFQFIEVGRKDPKKKLLRQRKREFVEIHDLFKPQQAADQAHRCLGCGNPYCEWKCPVHNFIPNWLKLVSEGNILAAAELSHQTNTLPEVCGRVCPQDRLCEGACTLNDGFGAVTIGSVEKYITDTAFAMGWRPDLSKVKPTGRRVAVIGAGPAGLGCADVLVRNGVTPVVFDRNPEIGGLLTFGIPEFKLEKHVLSRRREVFTGMGIEFRLNTEIGKDVTMEQLLDEYDAVFMGMGTYTYMKGGFPGEDLPGVHDALDFLIANVNRNLGFEKSAEDFVDMKGKRVVVLGGGDTAMDCNRTSIRQGAKSVTCAYRRDEENMPGSRKEVKNAKEEGVKFLFNRQPIAIVGEDRVEGVKVVETRLGEPDARGRRSPEPIPGSEEIIPAEAVLIAFGFRPSPAPWFERFEIATDSQGRVVAPAQAQFKHQTSNPKIFAGGDMVRGSDLVVTAIFEGRTAAEGILDYLGV |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA5035 and its homologs
|