| Identification |
| Name: |
4-diphosphocytidyl-2-C-methyl-D-erythritol kinase |
| Synonyms: |
- CMK
- 4-(cytidine-5'-diphospho)-2-C-methyl-D-erythritol kinase
|
| Gene Name: |
ispE |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in ATP binding |
| Specific Function: |
Catalyzes the phosphorylation of the position 2 hydroxy group of 4-diphosphocytidyl-2C-methyl-D-erythritol. Phosphorylates isopentenyl phosphate at low rates. Also acts on isopentenol, and, much less efficiently, dimethylallyl alcohol. Dimethylallyl monophosphate does not serve as a substrate |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol | + | 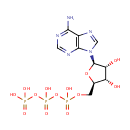 | + | 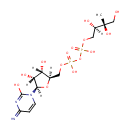 | → | Adenosine diphosphate | + |  | + | 2-phospho-4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol | + | 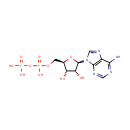 | + | 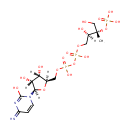 |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
| PAMDB000898 | 2-Phospho-4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol | MetaboCard | | PAMDB000931 | 4-(Cytidine 5'-diphospho)-2-C-methyl-D-erythritol | MetaboCard | | PAMDB000148 | Adenosine triphosphate | MetaboCard | | PAMDB000345 | ADP | MetaboCard | | PAMDB001633 | Hydrogen ion | MetaboCard |
|
| GO Classification: |
| Function |
|---|
| 4-(cytidine 5'-diphospho)-2-C-methyl-D-erythritol kinase activity | | adenyl nucleotide binding | | adenyl ribonucleotide binding | | ATP binding | | binding | | catalytic activity | | kinase activity | | nucleoside binding | | purine nucleoside binding | | transferase activity | | transferase activity, transferring phosphorus-containing groups | | Process |
|---|
| cellular lipid metabolic process | | cellular metabolic process | | isoprenoid metabolic process | | lipid metabolic process | | metabolic process | | phosphate metabolic process | | phosphorus metabolic process | | phosphorylation | | primary metabolic process | | terpenoid biosynthetic process | | terpenoid metabolic process |
|
| Gene Properties |
| Locus tag: |
PA4669 |
| Strand: |
+ |
| Entrez Gene ID: |
881380 |
| Accession: |
NP_253358.1 |
| GI: |
15599864 |
| Sequence start: |
5237393 |
| Sequence End: |
5238241 |
| Sequence Length: |
848 |
| Gene Sequence: |
>PA4669
ATGAGCGTGCGCCTCAGCCTGCCAGCCCCGGCCAAGCTCAATCTGTTCCTGCATATCCTCGGCCGCCGCGACGATGGCTATCACGAGTTGCAGACCCTGTTCCAATTCCTCGACCACGGCGACGAGCTGCATTTCGAAGCCCGCCAGGACGGCCAGGTACGCCTGCACACGGAAATCGCCGGGGTGCCGCACGACAGCAACCTGATCGTGCGCGCCGCGCGCGGGCTGCAGGAAGCCTCCGGCAGCCCGCAAGGCGTCGACATCTGGCTGGACAAGCGCTTGCCCATGGGGGGCGGCATCGGCGGCGGCAGCTCCGACGCCGCCACGACGCTCCTGGCCTTGAACCATCTCTGGCAACTCGGCTGGGACGAGGACCGCATCGCCGCCCTGGGCCTGCGCCTGGGCGCCGACGTGCCGGTCTTCACCCGCGGACGCGCAGCTTTCGCCGAGGGCGTGGGCGAAAAGCTCACCCCGGTGGACATCCCGGAGCCCTGGTATCTCGTCGTGGTTCCGCAAGTACTTGTCAGCACAGCAGAAATTTTTTCAGATCCACTGTTGACACGCGATTCTCCCGCCATTAAAGTGCGCACCGTTCTCGAGGGGGACAGCCGAAACGACTGTCAGCCGGTAGTCGAGAGGCGTTACCCAGAAGTTCGTAACGCTTTGATCTTGCTAAACAAATTTGTTTCAGCTAGATTAACCGGCACTGGAGGTTGTGTGTTTGGGAGCTTCCCAAACAAAGCTGAAGCTGATAAAGTCTCGGCCCTTCTTCCAGACCACCTTCAGAGGTTCGTCGCCAAAGGAAGCAACATCTCGATGTTGCACCGGAAGCTGGAAACTCTGGTCTGA |
| Protein Properties |
| Protein Residues: |
282 |
| Protein Molecular Weight: |
30.8 kDa |
| Protein Theoretical pI: |
6.61 |
| Hydropathicity (GRAVY score): |
-0.078 |
| Charge at pH 7 (predicted): |
-1.89 |
| Protein Sequence: |
>PA4669
MSVRLSLPAPAKLNLFLHILGRRDDGYHELQTLFQFLDHGDELHFEARQDGQVRLHTEIAGVPHDSNLIVRAARGLQEASGSPQGVDIWLDKRLPMGGGIGGGSSDAATTLLALNHLWQLGWDEDRIAALGLRLGADVPVFTRGRAAFAEGVGEKLTPVDIPEPWYLVVVPQVLVSTAEIFSDPLLTRDSPAIKVRTVLEGDSRNDCQPVVERRYPEVRNALILLNKFVSARLTGTGGCVFGSFPNKAEADKVSALLPDHLQRFVAKGSNISMLHRKLETLV |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA4669 and its homologs
|





