| Identification |
| Name: |
Bifunctional polymyxin resistance protein ArnA |
| Synonyms: |
- Polymyxin resistance protein PmrI
- UDP-4-amino-4-deoxy-L-arabinose formyltransferase
- ArnAFT
- UDP-L-Ara4N formyltransferase
- UDP-glucuronic acid oxidase, UDP-4-keto-hexauronic acid decarboxylating
- ArnADH
- UDP-GlcUA decarboxylase
- UDP-glucuronic acid dehydrogenase
|
| Gene Name: |
arnA |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in hydroxymethyl-, formyl- and related transferase activity |
| Specific Function: |
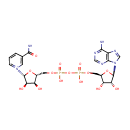
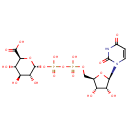
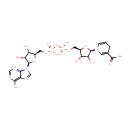
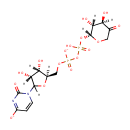
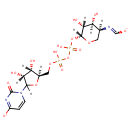
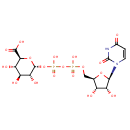
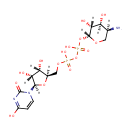
Bifunctional enzyme that catalyzes the oxidative decarboxylation of UDP-glucuronic acid (UDP-GlcUA) to UDP-4-keto- arabinose (UDP-Ara4O) and the addition of a formyl group to UDP-4- amino-4-deoxy-L-arabinose (UDP-L-Ara4N) to form UDP-L-4-formamido- arabinose (UDP-L-Ara4FN). The modified arabinose is attached to lipid A and is required for resistance to polymyxin and cationic antimicrobial peptides |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
- Amino sugar and nucleotide sugar metabolism pae00520
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
| | |
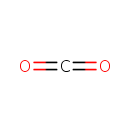
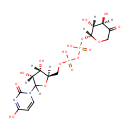
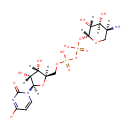 | + | an N10-formyl-tetrahydrofolate | + | 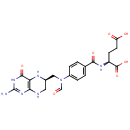 | → | 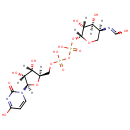 | + |  | + | a tetrahydrofolate | + |  |
| | | |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| binding | | catalytic activity | | coenzyme binding | | cofactor binding | | glycine hydroxymethyltransferase activity | | hydroxymethyl-, formyl- and related transferase activity | | methyltransferase activity | | transferase activity | | transferase activity, transferring one-carbon groups | | Process |
|---|
| biosynthetic process | | cellular metabolic process | | metabolic process |
|
| Gene Properties |
| Locus tag: |
PA3554 |
| Strand: |
+ |
| Entrez Gene ID: |
878473 |
| Accession: |
NP_252244.1 |
| GI: |
15598750 |
| Sequence start: |
3982021 |
| Sequence End: |
3984009 |
| Sequence Length: |
1988 |
| Gene Sequence: |
>PA3554
ATGACCTCGAAAGCCGTCGTCTTCGCCTACCACGACATCGGTTGCACCGGTATCGAAGCCCTGCTCAATGCCGGCTACGAGATCGCCGCCGTCTTCACCCATGCCGACGACCCACGGGAAAACACCTTCTACGCCTCGGTCGCACGCCTCTGCGCCGAGCGCGGCATTCCGCTGCACGCGCCCGAGGACGTGAACCATCCGCTGTGGCTGGAGCGTATCCGCCAACTGCGCCCGGACTTCCTGTTCTCCTTCTACTACCGCCGCCTGCTCGGCGCCGAGCTGCTCGCCTGCGCCGCACGCGGCGCCTACAACCTGCACGGTTCGCTGCTGCCGCGCTACCGCGGACGCGCCCCGGCGAACTGGGTGCTGGTCAACGGCGAAACGCAGACCGGGGTGACCCTGCATCGCATGATCGAGCGCGCCGACGCCGGGCCGATCCTCGCCCAGCAGGCCGTCGCCATCGACCCCGAGGACACCGCCCTGAGCCTGCACGGCAAGCTGCGCAAGGCCGCCGGCGCCCTGCTGCGCGACAGCCTGCCGCTGCTCGCCCTCGGCGTGCTGCCGGAAGTCGAGCAGGACGAGAGCCAGGCCAGCCACTTCGGCCGGCGCACCCCGGCGGACGGCCTGCTCGACTGGCACAGGCCGGCACGGCAGTTGTACGACCTGGTGCGCGCGGTGACCCAGCCCTACCCTGGCGCCTTCTGCCAGGTCGGCGAACAGAAGCTGATCGTCTGGAGCGCCGAGGTGGTCGCCGGCAACCACGGCCGCGAGCCGGGCAGCGTACTGTCCTGCGACCCGCTGCGGATCGCCTGCGGCGAGGACTCGCTGGTGCTGCGCTTCGGCCAGCGCGGCGAGCGCGGCCTGTACCTGGCCGGCACGCAACTGGCCACCGAGCTGGGCCTGGTCGAGGGCGCGCGCCTGCGTGGCGCGGCATGCAGTCCGCAGCGCCGCACGCGGGTGCTGATCCTCGGGGTCAACGGCTTCATCGGCAACCACCTGTCCGAACGCCTGCTGCGCGACGGTCGCTACGAGGTCCACGGCATGGACATCGGCTCCGACGCCATCGAACGGCTCAAGGCCGACCCGCATTTCCACTTCGTCGAAGGCGACATCGGCATCCATTCGGAGTGGCTCGAATACCATGTGAAGAAATGCGACGTGATCCTGCCGCTGGTGGCCATCGCCACGCCCATCGAGTACACGCGCAACCCGCTGCGGGTGTTCGAACTGGACTTCGAGGAAAACCTGCGGATCGTCCGCTACTGCGTGAAATACGGCAAACGCGTGGTGTTCCCTTCCACCTCCGAGGTCTACGGCATGTGCCAGGACCCGGACTTCGACGAAGACCGCTCGAACCTGGTGGTCGGGCCGATCAACAAGCAGCGCTGGATCTACTCGGTGTCCAAGCAGTTGCTCGACCGGGTGATCTGGGCCTACGGCCAGCAGGGCCTGCGCTTCACCCTGTTCCGTCCGTTCAACTGGATGGGCCCGCGCCTGGACCGCCTGGATTCGGCGCGGATCGGCAGCTCGCGGGCGATCACCCAGCTCATCCTGCACCTGGTCGAAGGCACGCCGATCCGCCTGGTCGACGGCGGCGCGCAGAAGCGCTGCTTCACCGACGTCGACGACGGCATCGAGGCCCTCGCGCGGATCATCGACAACCGCGACGGCCGCTGCGACGGGCAGATCGTCAACATCGGCAACCCGGACAACGAGGCGAGCATCCGCCAGCTCGGCGAGGAACTGCTGCGCCAGTTCGAGGCCCACCCGCTGCGCGCGCAGTTCCCGCCCTTCGCCGGCTTCCGCGAGGTGGAGAGCCGCAGCTTCTACGGCGACGGCTACCAGGACGTGGCCCACCGCAAGCCGAGCATCGACAACGCCCGGCGCCTGCTCGACTGGCAGCCCACCATCGAACTGCGCGAGACCATCGGCAAGACCCTCGACTTCTTCCTCCACGAAGCGCTCCGCGAGCGCGAGGCACAGGCGTGA |
| Protein Properties |
| Protein Residues: |
662 |
| Protein Molecular Weight: |
74.4 kDa |
| Protein Theoretical pI: |
6.68 |
| Hydropathicity (GRAVY score): |
-0.253 |
| Charge at pH 7 (predicted): |
-3.52 |
| Protein Sequence: |
>PA3554
MTSKAVVFAYHDIGCTGIEALLNAGYEIAAVFTHADDPRENTFYASVARLCAERGIPLHAPEDVNHPLWLERIRQLRPDFLFSFYYRRLLGAELLACAARGAYNLHGSLLPRYRGRAPANWVLVNGETQTGVTLHRMIERADAGPILAQQAVAIDPEDTALSLHGKLRKAAGALLRDSLPLLALGVLPEVEQDESQASHFGRRTPADGLLDWHRPARQLYDLVRAVTQPYPGAFCQVGEQKLIVWSAEVVAGNHGREPGSVLSCDPLRIACGEDSLVLRFGQRGERGLYLAGTQLATELGLVEGARLRGAACSPQRRTRVLILGVNGFIGNHLSERLLRDGRYEVHGMDIGSDAIERLKADPHFHFVEGDIGIHSEWLEYHVKKCDVILPLVAIATPIEYTRNPLRVFELDFEENLRIVRYCVKYGKRVVFPSTSEVYGMCQDPDFDEDRSNLVVGPINKQRWIYSVSKQLLDRVIWAYGQQGLRFTLFRPFNWMGPRLDRLDSARIGSSRAITQLILHLVEGTPIRLVDGGAQKRCFTDVDDGIEALARIIDNRDGRCDGQIVNIGNPDNEASIRQLGEELLRQFEAHPLRAQFPPFAGFREVESRSFYGDGYQDVAHRKPSIDNARRLLDWQPTIELRETIGKTLDFFLHEALREREAQA |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA3554 and its homologs
|