| Identification |
| Name: |
ATP-dependent RNA helicase rhlB |
| Synonyms: |
Not Available |
| Gene Name: |
rhlB |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Replication, recombination and repair |
| Specific Function: |
Can carry out ATP-dependent unwinding of double stranded RNA. Has a role in RNA decay. Involved in the RNA degradosome, a multi-enzyme complex important in RNA processing and messenger RNA degradation |
| Cellular Location: |
Cytoplasmic |
| KEGG Pathways: |
|
| KEGG Reactions: |
Not Available |
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
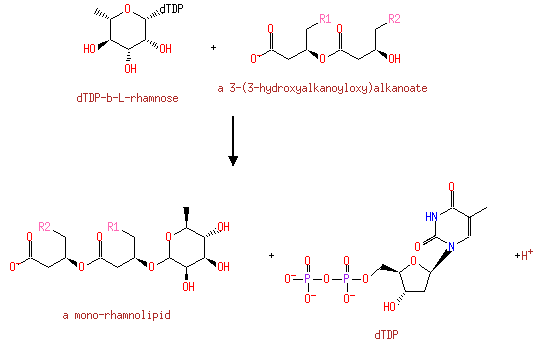
| | | | | | | | | | | | | | | dTDP-β-L-rhamnose + a 3-(3-hydroxyalkanoyloxy)alkanoate → a mono-rhamnolipid + dTDP + H +ReactionCard |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| adenyl nucleotide binding | | adenyl ribonucleotide binding | | ATP binding | | ATP-dependent helicase activity | | ATPase activity | | ATPase activity, coupled | | binding | | catalytic activity | | helicase activity | | hydrolase activity | | hydrolase activity, acting on acid anhydrides | | hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides | | nucleic acid binding | | nucleoside binding | | nucleoside-triphosphatase activity | | purine nucleoside binding | | pyrophosphatase activity |
|
| Gene Properties |
| Locus tag: |
PA3478 |
| Strand: |
- |
| Entrez Gene ID: |
878954 |
| Accession: |
NP_252168.1 |
| GI: |
15598674 |
| Sequence start: |
3890775 |
| Sequence End: |
3892055 |
| Sequence Length: |
1280 |
| Gene Sequence: |
>PA3478
ATGCACGCCATCCTCATCGCCATCGGCTCGGCCGGCGACGTATTTCCCTTCATCGGCCTGGCCCGGACCCTGAAACTGCGCGGGCACCGCGTGAGCCTCTGCACCATCCCGGTGTTTCGCGACGCGGTGGAGCAGCACGGCATCGCGTTCGTCCCGCTGAGCGACGAACTGACCTACCGCCGGACCATGGGCGATCCGCGCCTGTGGGACCCCAAGACGTCCTTCGGCGTGCTCTGGCAAGCCATCGCCGGGATGATCGAGCCGGTCTACGAGTACGTCTCGGCGCAGCGCCATGACGACATCGTGGTGGTCGGCTCGCTATGGGCGCTGGGCGCACGCATCGCTCACGAGAAGTACGGGATTCCCTACCTGTCCGCGCAGGTCTCGCCATCGACCCTGTTGTCGGCGCACCTGCCGCCGGTACACCCCAAGTTCAACGTGCCCGAGCAGATGCCGCTGGCGATGCGCAAGCTGCTCTGGCGCTGCATCGAGCGCTTCAAGCTGGATCGCACCTGCGCGCCGGAGATCAACGCGGTGCGCCGCAAGGTCGGCCTGGAAACGCCGGTGAAGCGCATCTTCACCCAATGGATGCATTCGCCGCAGGGCGTGGTCTGCCTGTTCCCGGCCTGGTTCGCGCCGCCCCAGCAGGATTGGCCGCAACCCCTGCACATGACCGGCTTCCCGCTGTTCGACGGCAGTATCCCGGGGACCCCGCTCGACGACGAACTGCAACGCTTTCTCGATCAGGGCAGCCGGCCGCTGGTGTTCACCCAGGGCTCGACCGAACACCTGCAGGGCGACTTCTACGCCATGGCCCTGCGCGCGCTGGAACGCCTCGGCGCGCGTGGGATCTTCCTCACCGGCGCCGGCCAGGAACCGCTGCGCGGCTTGCCGAACCACGTGCTGCAGCGCGCCTACGCGCCACTGGGAGCCTTGCTGCCATCGTGCGCCGGGCTGGTCCATCCGGGCGGTATCGGCGCCATGAGCCTAGCCTTGGCGGCGGGGGTGCCGCAGGTGCTGCTGCCCTGTGCCCACGACCAGTTCGACAATGCCGAACGGCTGGTCCGGCTCGGCTGCGGGATGCGCCTGGGCGTGCCGTTGCGCGAGCAGGAGTTGCGCGGGGCGCTGTGGCGCTTGCTCGAGGACCCGGCCATGGCGGCGGCCTGTCGGCGTTTCATGGAATTGTCACAACCGCACAGTATCGCTTGCGGTAAAGCGGCCCAGGTGGTCGAACGTTGTCATAGGGAGGGGGATGCTCGATGGCTGAAGGCTGCGTCCTGA |
| Protein Properties |
| Protein Residues: |
426 |
| Protein Molecular Weight: |
47.1 kDa |
| Protein Theoretical pI: |
8.57 |
| Hydropathicity (GRAVY score): |
0.014 |
| Charge at pH 7 (predicted): |
9.31 |
| Protein Sequence: |
>PA3478
MHAILIAIGSAGDVFPFIGLARTLKLRGHRVSLCTIPVFRDAVEQHGIAFVPLSDELTYRRTMGDPRLWDPKTSFGVLWQAIAGMIEPVYEYVSAQRHDDIVVVGSLWALGARIAHEKYGIPYLSAQVSPSTLLSAHLPPVHPKFNVPEQMPLAMRKLLWRCIERFKLDRTCAPEINAVRRKVGLETPVKRIFTQWMHSPQGVVCLFPAWFAPPQQDWPQPLHMTGFPLFDGSIPGTPLDDELQRFLDQGSRPLVFTQGSTEHLQGDFYAMALRALERLGARGIFLTGAGQEPLRGLPNHVLQRAYAPLGALLPSCAGLVHPGGIGAMSLALAAGVPQVLLPCAHDQFDNAERLVRLGCGMRLGVPLREQELRGALWRLLEDPAMAAACRRFMELSQPHSIACGKAAQVVERCHREGDARWLKAAS |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA3478 and its homologs
|