| Identification |
| Name: |
Siroheme synthase |
| Synonyms: |
- Uroporphyrinogen-III C-methyltransferase
- Urogen III methylase
- SUMT
- Uroporphyrinogen III methylase
- UROM
- Precorrin-2 dehydrogenase
- Sirohydrochlorin ferrochelatase
|
| Gene Name: |
cysG |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in methyltransferase activity |
| Specific Function: |
Multifunctional enzyme that catalyzes the SAM-dependent methylation of uroporphyrinogen III at position C-2 and C-7 to form precorrin-2 and then position C-12 or C-18 to form trimethylpyrrocorphin 2. It also catalyzes the conversion of precorrin-2 into siroheme. This reaction consists of the NAD- dependent oxidation of precorrin-2 into sirohydrochlorin and its subsequent ferrochelation into siroheme |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
 | + | S-adenosyl-L-methionine | → |  | + |  | + | 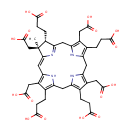 |
| | |
 | + | S-adenosyl-L-methionine | → | Precorrin-2 | + |  |
| | | | | | | |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| binding | | catalytic activity | | methyltransferase activity | | transferase activity | | transferase activity, transferring one-carbon groups | | Process |
|---|
| metabolic process | | nitrogen compound metabolic process | | oxidation reduction | | porphyrin biosynthetic process | | porphyrin metabolic process | | tetrapyrrole metabolic process |
|
| Gene Properties |
| Locus tag: |
PA2611 |
| Strand: |
- |
| Entrez Gene ID: |
882317 |
| Accession: |
NP_251301.1 |
| GI: |
15597807 |
| Sequence start: |
2952017 |
| Sequence End: |
2953414 |
| Sequence Length: |
1397 |
| Gene Sequence: |
>PA2611
ATGGATTTCCTGCCGCTGTTCCACTCCTTGCAAGGGCGCCTCGCGCTGGTGGTCGGCGGCGGCGAAGTCGCCCTGCGCAAGGCGCGCCTGCTGGCCGACGCCGGCGCGCGCCTGCGCGTGGTCGCGCCGCAGATCCACATCGAACTGCGGCATCTGGTGGAGCAGGGCGGCGGCGAGCTGCTCGAACGCGACTACCAGGACGGCGACCAGCCCGGCTGCGCCCTGATCATCGCCGCCACCGACGACGAACCGCTGAACGCCGAGGTGTCGCGAGCGGCCAATGCGCGCGGGATACCGGTCAACGTGGTGGACGCGCCGGCCCTGTGCAGCGTGATCTTCCCGGCCATCGTCGACCGTTCGCCGCTGGTGGTGGCGGTCTCCAGCGGCGGCGATGCGCCCGTGCTGGCGCGGCTGATCCGGGCCAAGCTGGAAACCTGGATTCCCTCGACCTACGGGCAACTGGCCGGACTCGCCAGCCGCTTCCGCCACCGGGTCAAGGAATTGCTACCCGACCTGCAGCAGCGCCGGGTGTTCTGGGAAAACCTGTTCCAGGGCGAGATCGCCGAGCGTGTCCTGGCCGGCCGTCCGGCGGAAGCCGAGCGTCTCCTGGAGGAGCACCTGGCCGGCGGCCTCGCGCACATAGCCACGGGCGAGGTATATCTGGTCGGGGCCGGGCCGGGGGATCCGGACCTGCTGACGTTCCGTGCCCTGCGCCTGATGCAGCAGGCCGACGTGGTGCTCTACGATCGCCTGGTGGCGCCGTCGATCCTCGAATTATGCCGCCGCGATGCCGAACGCCTGTACGTCGGCAAGCGCCGCGCCGAACATGCGGTGCCGCAGGATCGGATCAACCGCCTGCTGGTGGAGCTGGCCAGCCAGGGCAAGCGCGTGCTGCGCCTGAAGGGTGGCGACCCGTTCATCTTCGGCCGCGGCGGTGAGGAAATCGACGAACTGGCTGCCCACGGCATACCTTTCCAGGTCGTGCCGGGAATCACCGCGGCCAGCGGCTGCGCGGCCTACGCCGGGATTCCACTGACTCATCGCGATCATGCGCAATCGGTTCGTTTCGTCACCGGTCATCTGAAGGACGGCACCACCGACCTGCCGTGGCAGGACCTGGTAGCGCCTGGCCAGACCCTGGTGTTCTACATGGGCCTGGTGGGATTGCCGGTGATCTGCGAGCAACTGGTCGCCCACGGTCGCTCCGCACAGACCCCGGCGGCGCTGATCCAGCAGGGGACCACCGCCCAGCAGCGGGTATTCACCGGAACCCTGGAAAACCTGCCGCAACTGGTGGCTGAGCACGAAGTGCATGCGCCGACCCTGGTGATCGTCGGCGAAGTCGTGCAACTGCGCGACAAGCTGGCCTGGTTCGAAGGCGCCCGCGAGGACGCCTGA |
| Protein Properties |
| Protein Residues: |
465 |
| Protein Molecular Weight: |
50.4 kDa |
| Protein Theoretical pI: |
6.28 |
| Hydropathicity (GRAVY score): |
0.053 |
| Charge at pH 7 (predicted): |
-5.76 |
| Protein Sequence: |
>PA2611
MDFLPLFHSLQGRLALVVGGGEVALRKARLLADAGARLRVVAPQIHIELRHLVEQGGGELLERDYQDGDQPGCALIIAATDDEPLNAEVSRAANARGIPVNVVDAPALCSVIFPAIVDRSPLVVAVSSGGDAPVLARLIRAKLETWIPSTYGQLAGLASRFRHRVKELLPDLQQRRVFWENLFQGEIAERVLAGRPAEAERLLEEHLAGGLAHIATGEVYLVGAGPGDPDLLTFRALRLMQQADVVLYDRLVAPSILELCRRDAERLYVGKRRAEHAVPQDRINRLLVELASQGKRVLRLKGGDPFIFGRGGEEIDELAAHGIPFQVVPGITAASGCAAYAGIPLTHRDHAQSVRFVTGHLKDGTTDLPWQDLVAPGQTLVFYMGLVGLPVICEQLVAHGRSAQTPAALIQQGTTAQQRVFTGTLENLPQLVAEHEVHAPTLVIVGEVVQLRDKLAWFEGAREDA |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA2611 and its homologs
|