| Identification |
| Name: |
Thymidylate synthase |
| Synonyms: |
|
| Gene Name: |
thyA |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in thymidylate synthase activity |
| Specific Function: |
Provides the sole de novo source of dTMP for DNA biosynthesis. This protein also binds to its mRNA thus repressing its own translation |
| Cellular Location: |
Cytoplasm |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
| SMPDB Reactions: |
|
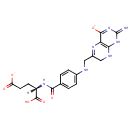 | + | 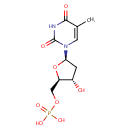 | + | Dihydrofolic acid | → | 5,10-Methylene-THF | + | 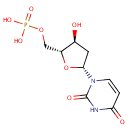 | + | 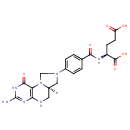 |
| | |
 | + | 5,10-methenyltetrahydrofolate mono-L-glutamate | + | 5,10-methenyltetrahydrofolate mono-L-glutamate | → |  | + |  | + | Dihydrofolic acid |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| 5,10-methylenetetrahydrofolate-dependent methyltransferase activity | | catalytic activity | | methyltransferase activity | | thymidylate synthase activity | | transferase activity | | transferase activity, transferring one-carbon groups | | Process |
|---|
| cellular nitrogen compound metabolic process | | dTMP biosynthetic process | | metabolic process | | nitrogen compound metabolic process | | nucleobase, nucleoside and nucleotide metabolic process | | nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | | nucleoside phosphate metabolic process | | nucleotide metabolic process | | pyrimidine deoxyribonucleoside monophosphate biosynthetic process | | pyrimidine nucleoside monophosphate biosynthetic process | | pyrimidine nucleotide biosynthetic process | | pyrimidine nucleotide metabolic process |
|
| Gene Properties |
| Locus tag: |
PA0342 |
| Strand: |
+ |
| Entrez Gene ID: |
879417 |
| Accession: |
NP_249033.1 |
| GI: |
15595539 |
| Sequence start: |
384733 |
| Sequence End: |
385527 |
| Sequence Length: |
794 |
| Gene Sequence: |
>PA0342
ATGAAACAGTACCTCGACCTGATGCGCCACGTGCGCGAGCACGGCACCTTCAAGAGCGACCGCACCGGCACCGGCACCTATAGCGTGTTCGGCCACCAGATGCGCTTCGACCTGGCCGCGGGCTTCCCCCTGGTGACCACCAAGAAGTGCCACCTCAAGTCGATCGTCCACGAGCTGCTGTGGTTCCTCAAGGGCTCCACCAATATCGCCTACCTCAAGGAGCACGGCGTCTCGATCTGGGACGAATGGGCCGACGAGAACGGCGACCTCGGCCCGGTGTATGGCTACCAGTGGCGCTCCTGGCCGGCGCCGGACGGCCGCCACATCGACCAGATCGCCAACCTGATGGCGATGCTGAAGAAGAATCCGGACTCGCGCCGGCTGATCGTCTCCGCCTGGAACCCGGCGCTGATCGACGAGATGGCCCTGCCGCCCTGCCACGCGCTGTTCCAGTTCTACGTCGCCGACGGCAAGCTCAGCTGCCAGCTCTACCAACGTTCGGCGGACATCTTCCTCGGCGTACCCTTCAATATCGCCAGCTATGCCCTGCTGACCCTGATGGTGGCGCAGGTCGCCGGCCTGCAGCCGGGCGAGTTCATCTGGACCGGCGGCGATTGCCACCTGTACGCCAACCACCTGGAGCAGGCCGACCTGCAGCTGACCCGCGAGCCGCTGCCGTTGCCGAGCATGAAGCTGAATCCCGAGGTGAAGGATCTGTTCGACTTTCGCTTCGAGGACTTCGAGCTGGTCGGCTACGAGGCCCATCCGCACATCAAGGCACCGGTCGCGGTCTGA |
| Protein Properties |
| Protein Residues: |
264 |
| Protein Molecular Weight: |
30 kDa |
| Protein Theoretical pI: |
6.51 |
| Hydropathicity (GRAVY score): |
-0.154 |
| Charge at pH 7 (predicted): |
-3.24 |
| Protein Sequence: |
>PA0342
MKQYLDLMRHVREHGTFKSDRTGTGTYSVFGHQMRFDLAAGFPLVTTKKCHLKSIVHELLWFLKGSTNIAYLKEHGVSIWDEWADENGDLGPVYGYQWRSWPAPDGRHIDQIANLMAMLKKNPDSRRLIVSAWNPALIDEMALPPCHALFQFYVADGKLSCQLYQRSADIFLGVPFNIASYALLTLMVAQVAGLQPGEFIWTGGDCHLYANHLEQADLQLTREPLPLPSMKLNPEVKDLFDFRFEDFELVGYEAHPHIKAPVAV |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA0342 and its homologs
|




