|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB001378 |
|---|
|
Identification |
|---|
| Name: |
L-Ala-gamma-D-Glu-Dap |
|---|
| Description: | L-ala-gamma-D-glu-DAP is a member of the chemical class known as Hybrid Peptides. These are compounds containing at least two different types of amino acids (alpha, beta, gamma, delta). L-alanyl-gamma-D-glutamyl-meso-diaminopimelate is related to MppA. MppA is a periplasmic binding protein in Pseudomonas aeruginosa essential for uptake of the cell wall murein tripeptide L-alanyl-gamma-D-glutamyl-meso-diaminopimelate. (PMID 10438753) |
|---|
|
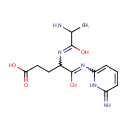
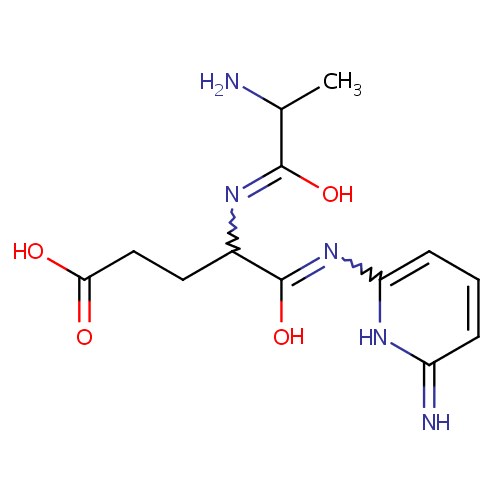
Structure |
|
|---|
| Synonyms: | - L-Ala-γ-D-Glu-diaminopimelate
- L-Ala-γ-D-glu-diaminopimelic acid
- L-Ala-g-D-glu-dap
- L-Ala-g-D-glu-diaminopimelate
- L-Ala-g-D-glu-diaminopimelic acid
- L-Ala-gamma-D-Glu-diaminopimelate
- L-Ala-gamma-D-Glu-diaminopimelic acid
- L-Ala-γ-D-glu-dap
- L-Ala-γ-D-glu-diaminopimelate
- L-Ala-γ-D-glu-diaminopimelic acid
- L-alanyl-γ-D-glutamyl-meso-diaminopimelate
- L-Alanyl-γ-D-glutamyl-meso-diaminopimelate
- L-Alanyl-γ-D-glutamyl-meso-diaminopimelic acid
- L-Alanyl-g-D-glutamyl-meso-diaminopimelate
- L-Alanyl-g-D-glutamyl-meso-diaminopimelic acid
- L-Alanyl-gamma-D-glutamyl-meso-diaminopimelate
- L-Alanyl-gamma-D-glutamyl-meso-diaminopimelic acid
- L-Alanyl-γ-D-glutamyl-meso-diaminopimelate
- L-Alanyl-γ-D-glutamyl-meso-diaminopimelic acid
|
|---|
|
Chemical Formula: |
C13H19N5O4 |
|---|
| Average Molecular Weight: |
309.3211 |
|---|
| Monoisotopic Molecular
Weight: |
309.143704121 |
|---|
| InChI Key: |
IBYVCSBRMYGDRR-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C13H19N5O4/c1-7(14)12(21)16-8(5-6-11(19)20)13(22)18-10-4-2-3-9(15)17-10/h2-4,7-8H,5-6,14H2,1H3,(H,16,21)(H,19,20)(H3,15,17,18,22) |
|---|
| CAS
number: |
Not Available |
|---|
| IUPAC Name: | 4-[(2-amino-1-hydroxypropylidene)amino]-4-[(6-imino-1,6-dihydropyridin-2-yl)-C-hydroxycarbonimidoyl]butanoic acid |
|---|
|
Traditional IUPAC Name: |
4-[(2-amino-1-hydroxypropylidene)amino]-4-[(6-imino-1H-pyridin-2-yl)-C-hydroxycarbonimidoyl]butanoic acid |
|---|
| SMILES: | CC(N)C(O)=NC(CCC(O)=O)C(O)=NC1=CC=CC(=N)N1 |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as peptides. These are compounds containing an amide derived from two or more amino carboxylic acid molecules (the same or different) by formation of a covalent bond from the carbonyl carbon of one to the nitrogen atom of another. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
|
Class |
Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
|
Direct Parent |
Peptides |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Alpha peptide
- Dihydropyridine
- Aminopyridine
- Imidolactam
- Pyridine
- Hydropyridine
- Heteroaromatic compound
- Azacycle
- Organoheterocyclic compound
- Organic 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Carboximidic acid derivative
- Carboximidic acid
- Hydrocarbon derivative
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Primary aliphatic amine
- Carbonyl group
- Amine
- Aromatic heteromonocyclic compound
|
|---|
| Molecular Framework |
Aromatic heteromonocyclic compounds |
|---|
| External Descriptors |
Not Available |
|---|
|
Physical Properties |
|---|
| State: |
Not Available |
|---|
| Charge: | -1 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Cytoplasm |
|---|
| Reactions: | |
|---|
|
Pathways: |
Not Available |
|---|
|
Spectra |
|---|
| Spectra: |
|
|---|
|
References |
|---|
| References: |
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Li, H., Park, J. T. (1999). "The periplasmic murein peptide-binding protein MppA is a negative regulator of multiple antibiotic resistance in Escherichia coli." J Bacteriol 181:4842-4847. Pubmed: 10438753
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
|
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|