|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB001302 |
|---|
|
Identification |
|---|
| Name: |
Ubiquinone-0 |
|---|
| Description: | Ubiquinone-1 is a member of the chemical class known as Benzoquinones. Normally in Pseudomonas aeruginosa the active form of Ubiquinone has 8 isoprene units (Ubiquinone-8) and in humans it normally has 10. Ubiquinone-10 is a naked version of Ubiquinone 8 that arises from conjugation by a shortened prenyl tail via 4-hydroxybenzoate polyprenyltransferase. Ubiquionone is involved in cellular respiration. It is fat-soluble and is therefore mobile in cellular membranes; it plays a unique role in the electron transport chain (ETC). In the inner bacterial membrane, electrons from NADH and succinate pass through the ETC to the oxygen, which is then reduced to water. The transfer of electrons through ETC results in the pumping of H+ across the membrane creating a proton gradient across the membrane, which is used by ATP synthase (located on the membrane) to generate ATP. |
|---|
|
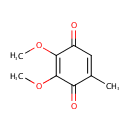
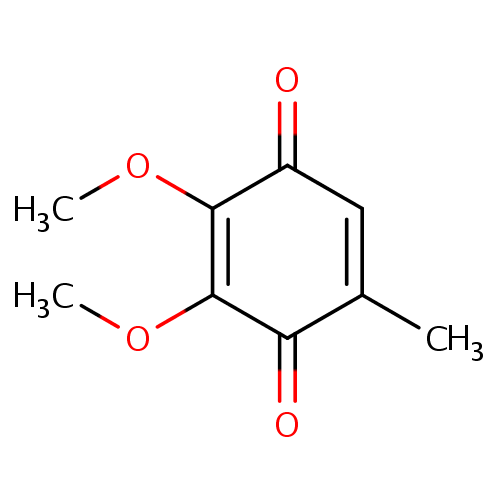
Structure |
|
|---|
| Synonyms: | - 2,3-Dimethoxy-5-methyl-1,4-benzoquinone
- 2,3-Dimethoxy-5-methyl-2,5-Cyclohexadiene-1,4-dione
- 2,3-Dimethoxy-5-methyl-p-benzoquinone
- 2,3-Dimethoxy-5-methylbenzo-1,4-quinone
- 2,3-Dimethoxy-5-methylbenzoquinone
- 2,3-Dimethoxy-5-methylcyclohexa-2,5-diene-1,4-dione
- 2,3-Dimethyoxyl-5-methyl-1,4-benzoquinone
- 2-Methyl-4,5-dimethoxy-p-quinone
- 2-Methyl-5,6-dimethoxybenzoquinone
- Coenzyme q0
- Coq0
- D9150_SIGMA
- Nchembio.62-comp10
- Q0
- Q0
- Ubiquinone 0
- Ubiquinone q0
- Ubiquinone-O
- Ubiquinone-Q0
- Ubiquinone-Q0
|
|---|
|
Chemical Formula: |
C9H10O4 |
|---|
| Average Molecular Weight: |
182.1733 |
|---|
| Monoisotopic Molecular
Weight: |
182.057908808 |
|---|
| InChI Key: |
UIXPTCZPFCVOQF-UHFFFAOYSA-N |
|---|
| InChI: | InChI=1S/C9H10O4/c1-5-4-6(10)8(12-2)9(13-3)7(5)11/h4H,1-3H3 |
|---|
| CAS
number: |
605-94-7 |
|---|
| IUPAC Name: | 2,3-dimethoxy-5-methylcyclohexa-2,5-diene-1,4-dione |
|---|
|
Traditional IUPAC Name: |
coenzyme Q0 |
|---|
| SMILES: | COC1=C(OC)C(=O)C(C)=CC1=O |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as ubiquinones. These are coenzyme Q derivatives containing a 5, 6-dimethoxy-3-methyl(1,4-benzoquinone) moiety to which an isoprenyl group is attached at ring position 2(or 6). |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
|
Class |
Prenol lipids |
|---|
| Sub Class | Quinone and hydroquinone lipids |
|---|
|
Direct Parent |
Ubiquinones |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Ubiquinone skeleton
- Quinone
- P-benzoquinone
- Vinylogous ester
- Cyclic ketone
- Ketone
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework |
Aliphatic homomonocyclic compounds |
|---|
| External Descriptors |
|
|---|
|
Physical Properties |
|---|
| State: |
Solid |
|---|
| Charge: | 0 |
|---|
|
Melting point: |
59-60 °C |
|---|
| Experimental Properties: |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Cytoplasm |
|---|
| Reactions: | |
|---|
|
Pathways: |
Not Available |
|---|
|
Spectra |
|---|
| Spectra: |
|
|---|
|
References |
|---|
| References: |
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
|
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|