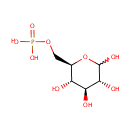
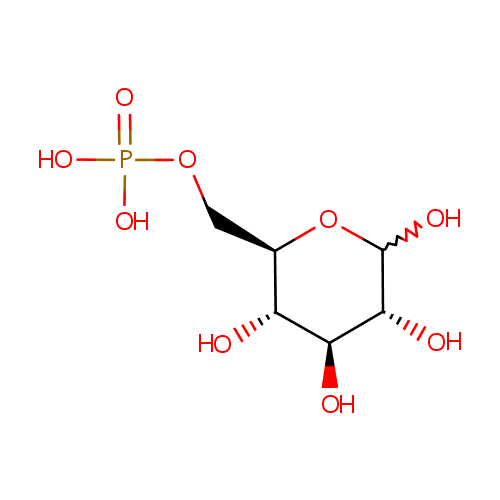
Glucose 6-phosphate (PAMDB000372)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000372 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Glucose 6-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Glucose 6 phosphate (alpha-D-glucose 6 phosphate or G6P) is the alpha-anomer of glucose-6-phosphate. There are two anomers of glucose 6 phosphate, the alpha anomer and the beta anomer Glucose-6-phosphate is a phosphorylated glucose molecule on carbon 6. When glucose enters a cell, it is immediately phosphorylated to G6P. This is catalyzed with hexokinase enzymes, thus consuming one ATP. A major reason for immediate phosphorylation of the glucose is so that it cannot diffuse out of the cell. The phosphorylation adds a charged group so the G6P cannot easily cross cell membranes. G6P can travel down two metabolic pathways, glycolysis and the pentose phosphate pathway. Note, the molecule now has 2 phosphoryl groups attached. The addition of the 2nd phosphoryl group is an irreversible step, so once this happens G6P will enter glycolysis and be turned into pyruvate (ATP production occurs). After being converted to G6P, phosphoglucose mutase (isomerase) can turn the molecule into glucose-1-phosphate. Glucose-1-phosphate can then be combined with uridine triphosphate (UTP) to form UDP-glucose. This reaction is driven by the hydrolysis of pyrophosphate that is released in the reaction. (Wikipedia) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C6H13O9P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 260.1358 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 260.029718526 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | NBSCHQHZLSJFNQ-GASJEMHNSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C6H13O9P/c7-3-2(1-14-16(11,12)13)15-6(10)5(9)4(3)8/h2-10H,1H2,(H2,11,12,13)/t2-,3-,4+,5-,6?/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 56-73-5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | {[(2R,3S,4S,5R)-3,4,5,6-tetrahydroxyoxan-2-yl]methoxy}phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | glucose 6-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OC1O[C@H](COP(O)(O)=O)[C@@H](O)[C@H](O)[C@H]1O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as monosaccharide phosphates. These are monosaccharides comprising a phosphated group linked to the carbohydrate unit. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Monosaccharides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Monosaccharide phosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Liquid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Phosphoenolpyruvic acid + D-Glucose > Glucose 6-phosphate + Pyruvic acid Glucose 1-phosphate <> Glucose 6-phosphate Glucose 6-phosphate + Water > D-Glucose + Phosphate Glucose 6-phosphate + NADP <> 6-Phosphonoglucono-D-lactone + Hydrogen ion + NADPH Glucose 6-phosphate + UDP-Glucose > Hydrogen ion + Trehalose 6-phosphate + Uridine 5'-diphosphate Adenosine triphosphate + D-Glucose > ADP + Glucose 6-phosphate + Hydrogen ion Arbutin 6-phosphate + Water > Glucose 6-phosphate + Hydroquinone Fructoselysine-6-phosphate + Water <> Glucose 6-phosphate + L-Lysine Glucose 6-phosphate <> Fructose 6-phosphate Water + Trehalose 6-phosphate > Glucose 6-phosphate + D-Glucose Adenosine triphosphate + alpha-D-Glucose <> ADP + Glucose 6-phosphate UDP-Glucose + Glucose 6-phosphate <> Uridine 5'-diphosphate + Trehalose 6-phosphate Protein N(pi)-phospho-L-histidine + D-Glucose <> Protein histidine + Glucose 6-phosphate Glucose 6-phosphate + Alpha-D-glucose 6-phosphate <> beta-D-Glucose 6-phosphate Phosphoenolpyruvic acid + b-D-Glucose > Glucose 6-phosphate + Pyruvic acid Cellobiose-6-phosphate + Water > Glucose 6-phosphate + b-D-Glucose beta-D-Glucose 1-phosphate Glucose 6-phosphate b-D-Glucose + Adenosine triphosphate > Hydrogen ion + Glucose 6-phosphate + ADP α-D-glucose 6-phosphate <> Glucose 6-phosphate Salicin 6-phosphate + Water > Glucose 6-phosphate + salicyl alcohol More...Trehalose 6-phosphate + Water > Glucose 6-phosphate + b-D-Glucose 6-Phospho-beta-D-glucosyl-(1,4)-D-glucose + Water > D-Glucose + Glucose 6-phosphate Glucose 6-phosphate + NADP > 6-Phosphonoglucono-D-lactone + NADPH Adenosine triphosphate + D-Glucose > ADP + Glucose 6-phosphate Cellobiose-6-phosphate + Water <> D-Glucose + Glucose 6-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in glucose-6-phosphate isomerase activity
- Specific function:
- D-glucose 6-phosphate = D-fructose 6- phosphate
- Gene Name:
- pgi
- Locus Tag:
- PA4732
- Molecular weight:
- 61.9 kDa
Reactions
| D-glucose 6-phosphate = D-fructose 6-phosphate. |
- General function:
- Involved in glucokinase activity
- Specific function:
- Not highly important in Pseudomonas aeruginosa as glucose is transported into the cell by the PTS system already as glucose 6-phosphate
- Gene Name:
- glk
- Locus Tag:
- PA3193
- Molecular weight:
- 34.6 kDa
Reactions
| ATP + D-glucose = ADP + D-glucose 6-phosphate. |
- General function:
- Involved in glucose-6-phosphate dehydrogenase activity
- Specific function:
- D-glucose 6-phosphate + NADP(+) = D-glucono- 1,5-lactone 6-phosphate + NADPH
- Gene Name:
- zwf
- Locus Tag:
- PA3183
- Molecular weight:
- 55.6 kDa
Reactions
| D-glucose 6-phosphate + NADP(+) = 6-phospho-D-glucono-1,5-lactone + NADPH. |
- General function:
- Involved in intramolecular transferase activity, phosphotransferases
- Specific function:
- This enzyme participates in both the breakdown and synthesis of glucose
- Gene Name:
- pgm
- Locus Tag:
- PA5131
- Molecular weight:
- 55.6 kDa
Reactions
| Alpha-D-glucose 1-phosphate = alpha-D-glucose 6-phosphate. |