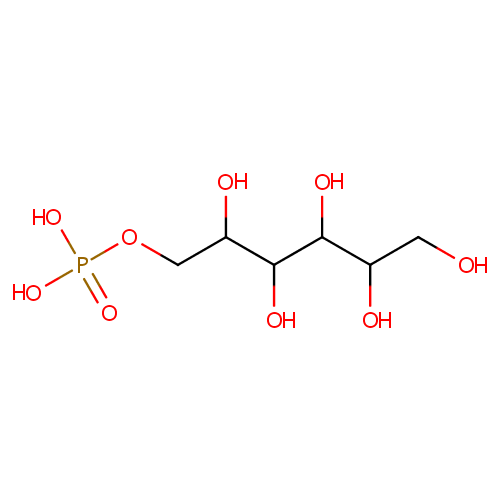
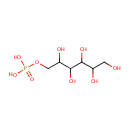Sorbitol-6-phosphate (PAMDB000371)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000371 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Sorbitol-6-phosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Sorbitol 6-phosphate (Sor6P) is an intermediate in sorbitol biosynthesis. It is a competitive inhibitor for both cytosolic and chloroplastic PGIs with a K(i) of 61 and 40muM, respectively. PMID: 18242768 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C6H15O9P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 262.1517 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 262.04536859 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | GACTWZZMVMUKNG-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C6H15O9P/c7-1-3(8)5(10)6(11)4(9)2-15-16(12,13)14/h3-11H,1-2H2,(H2,12,13,14) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 20479-58-7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | [(2,3,4,5,6-pentahydroxyhexyl)oxy]phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | (2,3,4,5,6-pentahydroxyhexyl)oxyphosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OCC(O)C(O)C(O)C(O)COP(O)(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as monoalkyl phosphates. These are organic compounds containing a phosphate group that is linked to exactly one alkyl chain. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organophosphorus compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Organic phosphoric acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Phosphate esters | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Monoalkyl phosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Phosphoenolpyruvic acid + Sorbitol > Pyruvic acid + Sorbitol-6-phosphate Phosphoenolpyruvic acid + Mannitol > Sorbitol-6-phosphate + Pyruvic acid Sorbitol-6-phosphate + NAD <> Fructose 6-phosphate + Hydrogen ion + NADH Sorbitol-6-phosphate + NAD <> beta-D-Fructose 6-phosphate + NADH + Hydrogen ion Protein N(pi)-phospho-L-histidine + Mannitol <> Protein histidine + Sorbitol-6-phosphate Protein N(pi)-phospho-L-histidine + Sorbitol <> Protein histidine + Sorbitol-6-phosphate Sorbitol-6-phosphate + NAD > Fructose 6-phosphate + NADH | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in oxidation-reduction process
- Specific function:
- D-mannitol 1-phosphate + NAD(+) = D-fructose 6-phosphate + NADH
- Gene Name:
- mtlD
- Locus Tag:
- PA2342
- Molecular weight:
- 54.3 kDa
Reactions
| D-mannitol 1-phosphate + NAD(+) = D-fructose 6-phosphate + NADH. |