| Identification |
| Name: |
Diaminopimelate epimerase |
| Synonyms: |
|
| Gene Name: |
dapF |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in diaminopimelate epimerase activity |
| Specific Function: |
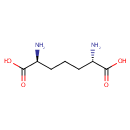
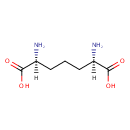
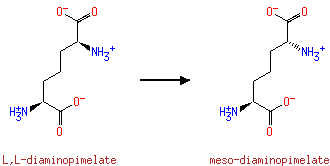
LL-2,6-diaminoheptanedioate = meso- diaminoheptanedioate |
| Cellular Location: |
Cytoplasm |
| KEGG Pathways: |
|
| Transports: | |
|---|
| Transport References: | - Orth, J. D., Conrad, T. M., Na, J., Lerman, J. A., Nam, H., Feist, A. M., Palsson, B. O. (2011). "A comprehensive genome-scale reconstruction of Escherichia coli metabolism--2011." Mol Syst Biol 7:535. Pubmed: 21988831
|
|---|
| KEGG Reactions: |
Not Available |
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
|
| Complex Reactions: |
Not Available |
| Transports: |
|
| Metabolites: |
| PAMDB ID | Name | View |
|---|
| PAMDB000360 | Diaminopimelic acid | MetaboCard | | PAMDB002055 | Meso-2,6-Diaminoheptanedioate | MetaboCard |
|
| GO Classification: |
| Component |
|---|
| cell part | | cytoplasm | | intracellular part | | Function |
|---|
| amino-acid racemase activity | | catalytic activity | | diaminopimelate epimerase activity | | isomerase activity | | racemase and epimerase activity | | racemase and epimerase activity, acting on amino acids and derivatives | | Process |
|---|
| aspartate family amino acid metabolic process | | cellular amino acid and derivative metabolic process | | cellular amino acid metabolic process | | cellular metabolic process | | lysine biosynthetic process | | lysine biosynthetic process via diaminopimelate | | lysine metabolic process | | metabolic process |
|
| Gene Properties |
| Locus tag: |
PA5278 |
| Strand: |
+ |
| Entrez Gene ID: |
877750 |
| Accession: |
NP_253965.1 |
| GI: |
15600471 |
| Sequence start: |
5942745 |
| Sequence End: |
5943575 |
| Sequence Length: |
830 |
| Gene Sequence: |
>PA5278
ATGCTTTTGCGCTTCACCAAGATGCATGGCCTGGGCAACGACTTCATGGTCCTCGACCTGGTCAGCCAGCACGCCCATGTGCAGCCCAAGCACGTCAAGCTCTGGGGCGACCGCAACACCGGAATCGGCTTCGACCAGTTGCTGATCGTCGAAGCCCCGAGCAGCCCCGACGTCGACTTCCGCTACCGGATCTTCAACGCCGACGGCTCCGAGGTCGAGCAGTGCGGCAACGGCGCTCGCTGCTTCGCCCGCTTCGTTCAGGACAAGCGCCTGACGGTGAAGAAGAGCATCCGCGTAGAAACCAAGGGCGGCATCATCGAGCTGAATATCCGCCCGGACGGCCAGGTCACGGTGGACATGGGTCCGCCGCGCCTGGCCCCGGCCGAGATACCGTTCCAGGCCGAGCGCGAAGCCTTGTCCTATGAGATAGAGGTGAATGGCCAGCGCGTCGAACTGGCCGCGGTATCCATGGGCAATCCCCATGGCGTGCTGCGCGTCGAGAACGTCGACAGCGCACCGGTACACAGCCTCGGTCCGCAACTGGAAGTCCATCCGCGCTTCCCGAAGAAGGCCAACATCGGCTTCCTCCAGGTGCTCGATCCGCACCATGCGCGCCTGCGCGTCTGGGAGCGCGGCGTCGGCGAGACCCAAGCCTGCGGCACCGGCGCCTGCGCCGCGGCGGTTGCCGGGATTCGCCAGGGCTGGCTGCAATCGCCGGTGCAGATCGACCTCCCCGGCGGCCGCCTGCACATCGAGTGGGCAGGTCCCGGCCAGCCCGTTATGATGACCGGGCCTGCCGTCCGCGTGTATGAAGGCCAGGTCCGCCTGTAA |
| Protein Properties |
| Protein Residues: |
276 |
| Protein Molecular Weight: |
30.3 kDa |
| Protein Theoretical pI: |
7.67 |
| Hydropathicity (GRAVY score): |
-0.212 |
| Charge at pH 7 (predicted): |
2.28 |
| Protein Sequence: |
>PA5278
MLLRFTKMHGLGNDFMVLDLVSQHAHVQPKHVKLWGDRNTGIGFDQLLIVEAPSSPDVDFRYRIFNADGSEVEQCGNGARCFARFVQDKRLTVKKSIRVETKGGIIELNIRPDGQVTVDMGPPRLAPAEIPFQAEREALSYEIEVNGQRVELAAVSMGNPHGVLRVENVDSAPVHSLGPQLEVHPRFPKKANIGFLQVLDPHHARLRVWERGVGETQACGTGACAAAVAGIRQGWLQSPVQIDLPGGRLHIEWAGPGQPVMMTGPAVRVYEGQVRL |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA5278 and its homologs
|