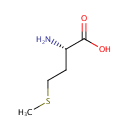| Identification |
| Name: |
Peptide methionine sulfoxide reductase msrA |
| Synonyms: |
- Protein-methionine-S-oxide reductase
- Peptide-methionine (S)-S-oxide reductase
- Peptide Met(O) reductase
|
| Gene Name: |
msrA |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor |
| Specific Function: |
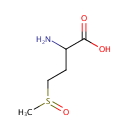
Could have an important function as a repair enzyme for proteins that have been inactivated by oxidation. Catalyzes the reversible oxidation-reduction of methionine sulfoxide in proteins to methionine |
| Cellular Location: |
Not Available |
| KEGG Pathways: |
Not Available |
| KEGG Reactions: |
|
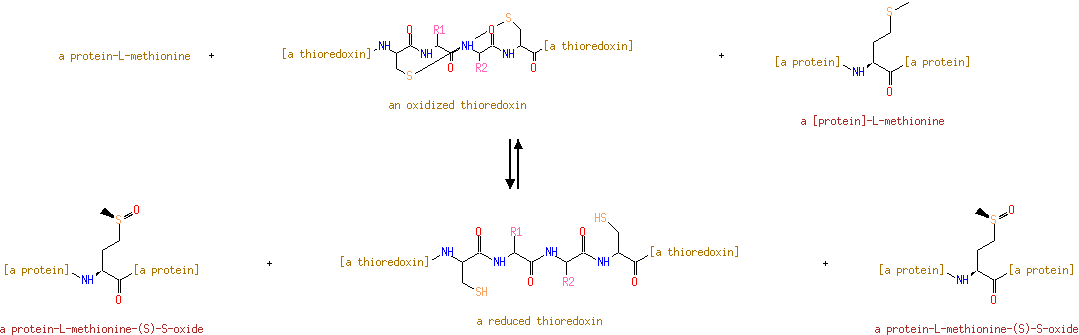
Peptide-L-methionine | + | Thioredoxin disulfide | + |  | + | 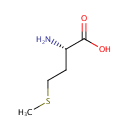 | ↔ | Peptide-L-methionine (S)-S-oxide | + | 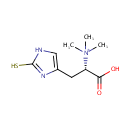 | + | 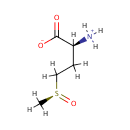 |
| |
|
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
|
Peptide-L-methionine | + | thioredoxin disulfide | + |  | → | peptide-L-methionine (S)-S-oxide | + | thioredoxin |
| Peptide-L-methionine + thioredoxin disulfide + Water → peptide-L-methionine (S)-S-oxide + thioredoxin ReactionCard | |
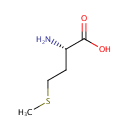 | + | thioredoxin disulfide | + |  | → |  | + | thioredoxin |
| | |
Peptide-L-methionine | + | Thioredoxin disulfide | + |  | + | 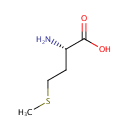 | ↔ | Peptide-L-methionine (S)-S-oxide | + |  | + |  |
| | | a protein-L-methionine + an oxidized thioredoxin + a [protein]-L-methionine = a protein-L-methionine-( S)- S-oxide + a reduced thioredoxin + a protein-L-methionine-( S)- S-oxide ReactionCard | |
Protein-L-methionine | + |  | → | Protein-L-methionine-R-S-oxides |
| | |
Protein-L-methionine | + |  | → | Protein-L-Methionine-S-Oxides |
| |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Function |
|---|
| catalytic activity | | oxidoreductase activity | | oxidoreductase activity, acting on a sulfur group of donors | | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor | | Process |
|---|
| macromolecule metabolic process | | metabolic process | | oxidation reduction | | protein metabolic process |
|
| Gene Properties |
| Locus tag: |
PA5018 |
| Strand: |
+ |
| Entrez Gene ID: |
881240 |
| Accession: |
NP_253705.1 |
| GI: |
15600211 |
| Sequence start: |
5643803 |
| Sequence End: |
5644450 |
| Sequence Length: |
647 |
| Gene Sequence: |
>PA5018
ATGGTTCTGCGTTCTGAAATCCTCACCAAGAAAAGCGAGCTGCCCACGCCGGACCAGGCCCTGCCGGGTCGCGAAAGCGCCATGCCGGTGCCCGAAGCGCACTTCGTCAACGGCCGCCCGCTGACCGCGCCGTTCCCCGCCGGCCTGCAACAGGTGCTGTTCGGCATGGGCTGCTTCTGGGGCGCCGAGCGTCGCCTCTGGCAGCAACCGGGTGTCTGGGTGACCGCGGTGGGTTATGCCGGCGGCTACACGCCGAACCCGACCTACGACGAAGTCTGCTCCGGCCTGACCGGCCACAGCGAGGTCGTGCTGGTGGTCTACAACCCGCAGGAGACCAGCTTCGAACAGCTGCTCAAGGTATTCTGGGAAGCCCACGATCCGACCCAGGGCATGCGCCAGGGCGGCGACATCGGCACCCAGTACCGCTCGGTGATCTACACCTTCGACGCGGCCCAGAAAGCCGCGGCCATGGCCAGCCGGGAGAGCTTCCAGGCCGAACTGGCGAAAGCCGGCTACGACCGCATCACCACCGAGATCGCCGACGTCCCGCCCTTCTACTACGCCGAGGCCTACCACCAGCAGTACCTGGCGAAGAACCCCAACGGCTACTGCGGCCTGGGCGGCACCGGGGTGTGCCTGCCGGCCTGA |
| Protein Properties |
| Protein Residues: |
215 |
| Protein Molecular Weight: |
23.5 kDa |
| Protein Theoretical pI: |
5 |
| Hydropathicity (GRAVY score): |
-0.243 |
| Charge at pH 7 (predicted): |
-5.17 |
| Protein Sequence: |
>PA5018
MVLRSEILTKKSELPTPDQALPGRESAMPVPEAHFVNGRPLTAPFPAGLQQVLFGMGCFWGAERRLWQQPGVWVTAVGYAGGYTPNPTYDEVCSGLTGHSEVVLVVYNPQETSFEQLLKVFWEAHDPTQGMRQGGDIGTQYRSVIYTFDAAQKAAAMASRESFQAELAKAGYDRITTEIADVPPFYYAEAYHQQYLAKNPNGYCGLGGTGVCLPA |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA5018 and its homologs
|