| Identification |
| Name: |
Probable 4-deoxy-4-formamido-L-arabinose-phosphoundecaprenol deformylase ArnD |
| Synonyms: |
Not Available |
| Gene Name: |
arnD |
| Enzyme Class: |
Not Available |
| Biological Properties |
| General Function: |
Involved in hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amides |
| Specific Function: |
Catalyzes the deformylation of 4-deoxy-4-formamido-L- arabinose-phosphoundecaprenol to 4-amino-4-deoxy-L-arabinose- phosphoundecaprenol. The modified arabinose is attached to lipid A and is required for resistance to polymyxin and cationic antimicrobial peptides (Probable) |
| Cellular Location: |
Cytoplasmic |
| KEGG Pathways: |
- Amino sugar and nucleotide sugar metabolism pae00520
|
| KEGG Reactions: |
Not Available |
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
| | |
4-deoxy-4-formamido-beta-L-arabinose di-trans,poly-cis-undecaprenyl phosphate | + |  | → | 4-amino-4-deoxy-alpha-L-arabinose di-trans,poly-cis-undecaprenyl phosphate | + | 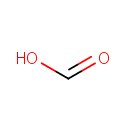 |
| 4-deoxy-4-formamido-beta-L-arabinose di-trans,poly-cis-undecaprenyl phosphate + Water → 4-amino-4-deoxy-alpha-L-arabinose di-trans,poly-cis-undecaprenyl phosphate + Formic acidReactionCard |
|
| Complex Reactions: |
| | |
4-deoxy-4-formamido-beta-L-arabinose di-trans,poly-cis-undecaprenyl phosphate | + |  | → | 4-amino-4-deoxy-alpha-L-arabinose di-trans,poly-cis-undecaprenyl phosphate | + |  |
| 4-deoxy-4-formamido-beta-L-arabinose di-trans,poly-cis-undecaprenyl phosphate + Water → 4-amino-4-deoxy-alpha-L-arabinose di-trans,poly-cis-undecaprenyl phosphate + Formic acidReactionCard |
|
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
| PAMDB000053 | Formic acid | MetaboCard | | PAMDB001760 | undecaprenyl phosphate-4-amino-4-deoxy-L-arabinose | MetaboCard | | PAMDB001761 | Undecaprenyl phosphate-4-amino-4-formyl-L-arabinose | MetaboCard | | PAMDB000142 | Water | MetaboCard |
|
| GO Classification: |
| Function |
|---|
| catalytic activity | | hydrolase activity | | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds | | Process |
|---|
| carbohydrate metabolic process | | metabolic process | | primary metabolic process |
|
| Gene Properties |
| Locus tag: |
PA3555 |
| Strand: |
+ |
| Entrez Gene ID: |
879121 |
| Accession: |
NP_252245.1 |
| GI: |
15598751 |
| Sequence start: |
3984006 |
| Sequence End: |
3984893 |
| Sequence Length: |
887 |
| Gene Sequence: |
>PA3555
GTGATCCAGGCCGGCCTGCGCATCGACGTCGACACCTTCCGCGGCACCCGCGACGGGGTGCCGCGGCTGCTCGAGCTGCTCGACGAAGCCGGGCTGAAGGCGACCTTCTTCTTCAGCGTCGGCCCGGACAACATGGGCCGCCACCTCTGGCGCCTGGCGCGCCCGGCGTTCTTCTGGAAGATGCTCCGCTCGCGCGCCGCCAGCCTGTATGGCTGGGACATCCTGCTGGCCGGCACCGCCTGGCCGGGCAAGCCGATCGGCCGCGAACTCGGCCCGCTGATGCGGCGGACCCAGGCCGCCGGCCACGAGGTCGGCCTGCACGCCTGGGACCACCATGCCTGGCAGACCCACGCCGGAGTCTGGAGCGTGCAGCAGCTCGGCGAGCAGATCCGCCGCGGCAGCGATTGCCTGGCGGACATCCTCGGCCAGCCGGTGCGTTGTTCCGCCGCCGCCGGCTGGCGCGCCGACGGCCGGGTGGTCGAGGCCAAGCAGCCCTTCGGCTTCCGCTACAACAGCGACTGCCGCGGACGCGGCGCGTTCCGCCCGCGCCTGGCCGACGGCAGCCCTGGCATCCCGCAGGTGCCGGTGAACCTGCCGACCTTCGACGAGGTGGTCGGTCCCGGCCTGCCGCGCGAGGCCTACAACGACTTCATCCTGGAACGCTTCGCCGCTGGCCGCGACAACGTCTACACCATCCACGCCGAGGTCGAGGGGCTGCTCCTCGCCCCGGCGTTCCGCGAACTGCTGCGGCGCGCCGAACGGCGCGGCATCCGCTTCCGACCGTTGGGCGAACTGCTGCCGGACGATCCGCGCAGCCTGCCGCTGGCCGAACTGGTGCGCGGCCGCCTGGCCGGCCGCGAGGGCTGGCTGGGAGTGCGCCAGCCATGA |
| Protein Properties |
| Protein Residues: |
295 |
| Protein Molecular Weight: |
32.9 kDa |
| Protein Theoretical pI: |
10.39 |
| Hydropathicity (GRAVY score): |
-0.285 |
| Charge at pH 7 (predicted): |
9.6 |
| Protein Sequence: |
>PA3555
MIQAGLRIDVDTFRGTRDGVPRLLELLDEAGLKATFFFSVGPDNMGRHLWRLARPAFFWKMLRSRAASLYGWDILLAGTAWPGKPIGRELGPLMRRTQAAGHEVGLHAWDHHAWQTHAGVWSVQQLGEQIRRGSDCLADILGQPVRCSAAAGWRADGRVVEAKQPFGFRYNSDCRGRGAFRPRLADGSPGIPQVPVNLPTFDEVVGPGLPREAYNDFILERFAAGRDNVYTIHAEVEGLLLAPAFRELLRRAERRGIRFRPLGELLPDDPRSLPLAELVRGRLAGREGWLGVRQP |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA3555 and its homologs
|