| Identification |
| Name: |
Undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase |
| Synonyms: |
- Polymyxin resistance protein pmrF
- Undecaprenyl-phosphate Ara4FN transferase
- Ara4FN transferase
|
| Gene Name: |
arnC |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
beta-L-Ara4N-lipid A biosynthetic process |
| Specific Function: |
phosphotransferase activity, for other substituted phosphate groups, transferase activity, transferring glycosyl groups |
| Cellular Location: |
Cell inner membrane; Multi-pass membrane protein |
| KEGG Pathways: |
- Amino sugar and nucleotide sugar metabolism pae00520
|
| KEGG Reactions: |
|
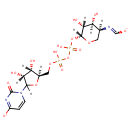 | + |  | ↔ | Undecaprenyl phosphate alpha-L-Ara4FN | + | 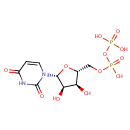 | + |  |
| |
|
| SMPDB Reactions: |
|
 | + | di-trans,octa-cis-undecaprenyl phosphate | → |  | + |  |
| |
|
| PseudoCyc/BioCyc Reactions: |
|
 | + |  | → | 4-deoxy-4-formamido-α-L-arabinopyranosyl ditrans,octacis-undecaprenyl phosphate | + |  |
| |
|
| Complex Reactions: |
|
| Transports: |
Not Available |
| Metabolites: |
| PAMDB ID | Name | View |
|---|
| PAMDB003490 | 4-Amino-4-deoxy-alpha-L-arabinopyranosyl di-trans,octa-cis-undecaprenyl phosphate | MetaboCard | | PAMDB006358 | 4-deoxy-4-formamido-α-L-arabinopyranosyl ditrans,octacis-undecaprenyl phosphate | MetaboCard | | PAMDB001145 | Di-trans,poly-cis-undecaprenyl phosphate | MetaboCard | | PAMDB003589 | UDP-4-Deoxy-4-formamido-beta-L-arabinose | MetaboCard | | PAMDB001054 | Undecaprenyl phosphate | MetaboCard | | PAMDB001761 | Undecaprenyl phosphate-4-amino-4-formyl-L-arabinose | MetaboCard | | PAMDB001763 | Uridine 5''-diphospho-{beta}-4-deoxy-4-formamido-L-arabinose | MetaboCard | | PAMDB000123 | Uridine 5'-diphosphate | MetaboCard |
|
| GO Classification: |
| Function |
|---|
| phosphotransferase activity, for other substituted phosphate groups | | transferase activity, transferring glycosyl groups | | Process |
|---|
| beta-L-Ara4N-lipid A biosynthetic process |
|
| Gene Properties |
| Locus tag: |
PA3553 |
| Strand: |
+ |
| Entrez Gene ID: |
878472 |
| Accession: |
NP_252243.1 |
| GI: |
15598749 |
| Sequence start: |
3981005 |
| Sequence End: |
3982024 |
| Sequence Length: |
1019 |
| Gene Sequence: |
>PA3553
ATGAAGCCCTATCCGATCGACCTGGTCTCGGTGGTGATCCCGGTCTACAACGAGGAAGCCAGCCTGCCCGAACTGCTGCGCCGCACCGAGGCCGCCTGCCTGGAGCTGGGCCGCGCCTTCGAGATCGTGCTGGTCGACGACGGCAGCCGCGACCGCTCCGCGGAACTCTTGCAGGCCGCCGCCGAACGCGACGGCAGCGCCGTGGTGGCGGTGATCCTCAACCGCAACTACGGCCAGCACGCGGCGATCCTCGCCGGCTTCGAACAGTCCCGTGGAGACCTGGTGATCACCCTCGATGCCGACCTGCAGAACCCGCCGGAGGAAATCCCGCGCCTGGTGGAGCGCGCCGCGCAGGGCTACGACGTGGTCGGCAGCATCCGCGCCGAGCGCCAGGACTCGGCCTGGCGGCGCTGGCCTTCGCGCCTGGTGAACCTGGCGGTGCAGCGCTCCACCGGGGTCGCCATGCACGACTACGGCTGCATGCTGCGCGCCTACCGGCGCAGCATCGTCGAAGCGATGCTGGCCTGCCGCGAACGCAGTACCTTCATCCCGATCCTGGCCAACGGCTTCGCCCGGCATACCTGCGAGATCCGCGTCGCCCACGCCGAGCGCGCCCACGGCGAATCGAAATACAGCGCGATGCGCCTGCTCAACCTGATGTTCGACCTGGTCACCTGCATGACCACCACGCCGCTGCGCCTGCTCAGCCTGGTCGGCGGCGGCATGGCCCTGGCCGGCTTCCTCTTCGCCCTGTTCCTCCTCGTGCTGCGCCTGGCCTTCGGCGCCGCCTGGGCCGGCAACGGCCTGTTCGTGCTGTTCGCCGTGCTGTTCATGTTCAGCGGCGTGCAGTTGCTCGGCATGGGCCTGCTCGGCGAGTACCTCGGGCGCATGTACAGCGACGTGCGCGCGCGGCCGCGCTTCTTCATCGAGCGCGTGGTCCGCGCAACGCCCTCCGCGCTTCCCTCGGCCCTGCAGCGGGCCGGTTTCACCTCTTCATCCAGCGAGCCTTCCACGCCATGA |
| Protein Properties |
| Protein Residues: |
339 |
| Protein Molecular Weight: |
37.3 kDa |
| Protein Theoretical pI: |
8.21 |
| Hydropathicity (GRAVY score): |
0.193 |
| Charge at pH 7 (predicted): |
3.05 |
| Protein Sequence: |
>PA3553
MKPYPIDLVSVVIPVYNEEASLPELLRRTEAACLELGRAFEIVLVDDGSRDRSAELLQAAAERDGSAVVAVILNRNYGQHAAILAGFEQSRGDLVITLDADLQNPPEEIPRLVERAAQGYDVVGSIRAERQDSAWRRWPSRLVNLAVQRSTGVAMHDYGCMLRAYRRSIVEAMLACRERSTFIPILANGFARHTCEIRVAHAERAHGESKYSAMRLLNLMFDLVTCMTTTPLRLLSLVGGGMALAGFLFALFLLVLRLAFGAAWAGNGLFVLFAVLFMFSGVQLLGMGLLGEYLGRMYSDVRARPRFFIERVVRATPSALPSALQRAGFTSSSSEPSTP |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA3553 and its homologs
|