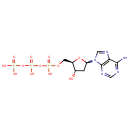| Identification |
| Name: |
DNA polymerase III subunit delta' |
| Synonyms: |
Not Available |
| Gene Name: |
holB |
| Enzyme Class: |
|
| Biological Properties |
| General Function: |
Involved in DNA binding |
| Specific Function: |
DNA polymerase III is a complex, multichain enzyme responsible for most of the replicative synthesis in bacteria. This DNA polymerase also exhibits 3' to 5' exonuclease activity |
| Cellular Location: |
Cytoplasmic |
| KEGG Pathways: |
|
| KEGG Reactions: |
|
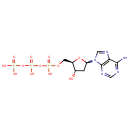 | + | DNA | ↔ | 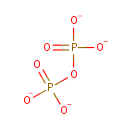 | + | DNA |
| | |
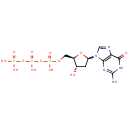 | + | DNA | ↔ |  | + | DNA |
| | |
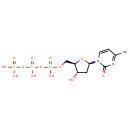 | + | DNA | ↔ |  | + | DNA |
| | |
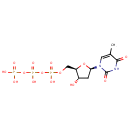 | + | DNA | ↔ |  | + | DNA |
| | |
Deoxynucleoside triphosphate | + | DNA | ↔ |  |
| |
|
| SMPDB Reactions: |
Not Available |
| PseudoCyc/BioCyc Reactions: |
|
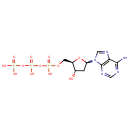 | + | DNA | ↔ |  | + | DNA |
| | |
 | + | DNA | ↔ |  | + | DNA |
| | |
 | + | DNA | ↔ |  | + | DNA |
| | |
 | + | DNA | ↔ |  | + | DNA |
| | |
Deoxynucleoside triphosphate | + | DNA(n) | → |  | + | DNA(n+1) |
| | |
Deoxynucleoside triphosphate | + | DNA | ↔ |  |
| |
|
| Complex Reactions: |
|
Deoxynucleoside triphosphate | + | DNA(n) | → |  | + | DNA(n+1) |
| |
|
| Transports: |
Not Available |
| Metabolites: |
|
| GO Classification: |
| Component |
|---|
| DNA polymerase complex | | DNA polymerase III complex | | macromolecular complex | | protein complex | | Function |
|---|
| 3'-5' exonuclease activity | | binding | | catalytic activity | | DNA binding | | DNA polymerase activity | | DNA-directed DNA polymerase activity | | exonuclease activity | | hydrolase activity | | hydrolase activity, acting on ester bonds | | nuclease activity | | nucleic acid binding | | nucleotidyltransferase activity | | transferase activity | | transferase activity, transferring phosphorus-containing groups | | Process |
|---|
| cellular macromolecule metabolic process | | DNA metabolic process | | DNA replication | | macromolecule metabolic process | | metabolic process |
|
| Gene Properties |
| Locus tag: |
PA2961 |
| Strand: |
- |
| Entrez Gene ID: |
880379 |
| Accession: |
NP_251651.1 |
| GI: |
15598157 |
| Sequence start: |
3320064 |
| Sequence End: |
3321050 |
| Sequence Length: |
986 |
| Gene Sequence: |
>PA2961
ATGGCTGATATCTATCCCTGGCAGCAGGCGCTCTGGAGCCAACTGGGCGGCCGCGCCCAGCACGCCCACGCCTATCTGCTCTACGGTCCCGCCGGGATCGGTAAGCGTGCCCTGGCCGAGCATTGGGCCGCCCAACTGCTCTGCCAGCGGCCTGCCGCCGCTGGCGCATGCGGCGAGTGCAAGGCCTGCCAGTTGCTGGCCGCCGGTACCCATCCGGACTACTTCGTGCTGGAGCCGGAGGAGGCGGAGAAACCGATCCGGGTCGATCAGGTCCGCGACCTGGTTGGCTTCGTGGTGCAGACCGCGCAACTGGGCGGGCGCAAGGTCGTGTTGCTCGAACCCGCCGAGGCGATGAACGTGAACGCCGCCAACGCCCTGTTGAAAAGCCTTGAAGAGCCGTCGGGCGATACTGTGCTGCTGCTGATCAGCCACCAGCCCAGCCGCCTGTTGCCGACCATCAAGAGCCGCTGCGTGCAGCAGGCCTGCCCTTTGCCGGGCGCGGCGGCGAGTCTGGAATGGCTGGCGCGGGCCCTGCCCGACGAGCCGGCCGAGGCCCTGGAGGAACTGTTGGCGCTTTCCGGCGGCTCGCCGCTCACCGCCCAGCGCCTGCACGGCCAGGGCGTGCGCGAGCAGCGCGCGCAGGTGGTCGAGGGGGTGAAGAAGCTGCTCAAGCAGCAGATCGCCGCCAGCCCACTGGCCGAAAGCTGGAACAGCGTGCCGTTGCCATTGCTCTTCGACTGGTTCTGCGACTGGACGCTGGGCATCCTGCGCTACCAGTTGACCCACGACGAGGAGGGGCTGGGGTTGGCCGACATGCGCAAGGTGATCCAGTACCTGGGCGACAAATCCGGGCAGGCCAAGGTGCTGGCCATGCAGGACTGGCTGCTGCAACAGCGGCAGAAAGTGTTGAACAAGGCCAACCTCAACCGTGTCCTGCTTCTCGAAGCCCTGCTGGTGCAGTGGGCAAGCCTGCCCGGGCCGGGCTAG |
| Protein Properties |
| Protein Residues: |
328 |
| Protein Molecular Weight: |
35.7 kDa |
| Protein Theoretical pI: |
6.62 |
| Hydropathicity (GRAVY score): |
-0.027 |
| Charge at pH 7 (predicted): |
-1.53 |
| Protein Sequence: |
>PA2961
MADIYPWQQALWSQLGGRAQHAHAYLLYGPAGIGKRALAEHWAAQLLCQRPAAAGACGECKACQLLAAGTHPDYFVLEPEEAEKPIRVDQVRDLVGFVVQTAQLGGRKVVLLEPAEAMNVNAANALLKSLEEPSGDTVLLLISHQPSRLLPTIKSRCVQQACPLPGAAASLEWLARALPDEPAEALEELLALSGGSPLTAQRLHGQGVREQRAQVVEGVKKLLKQQIAASPLAESWNSVPLPLLFDWFCDWTLGILRYQLTHDEEGLGLADMRKVIQYLGDKSGQAKVLAMQDWLLQQRQKVLNKANLNRVLLLEALLVQWASLPGPG |
| References |
| External Links: |
|
| General Reference: |
PaperBLAST - Find papers about PA2961 and its homologs
|