|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB110749 |
|---|
|
Identification |
|---|
| Name: |
(S)-malate |
|---|
| Description: | An optically active form of malate having (S)-configuration. |
|---|
|
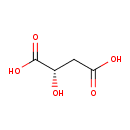
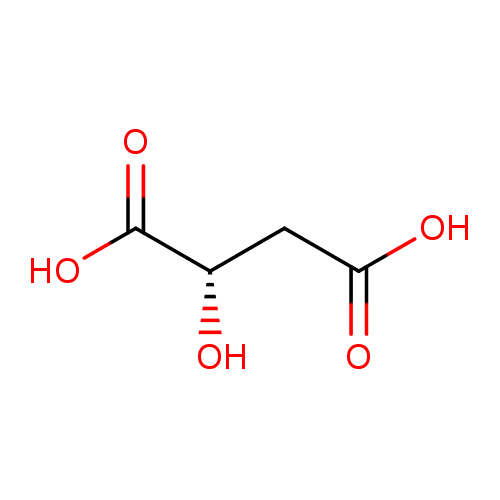
Structure |
|
|---|
| Synonyms: | -
(S)-malic acid
-
L-apple acid
-
L-malic acid
-
L-hydroxysuccinic acid
-
L-hydroxybutanedioic acid
-
L-malate
|
|---|
|
Chemical Formula: |
C4H4O5
|
|---|
| Average Molecular Weight: |
132.07 |
|---|
| Monoisotopic Molecular
Weight: |
134.0215233031 |
|---|
| InChI Key: |
BJEPYKJPYRNKOW-REOHCLBHSA-L |
|---|
| InChI: |
InChI=1S/C4H6O5/c5-2(4(8)9)1-3(6)7/h2,5H,1H2,(H,6,7)(H,8,9)/p-2/t2-/m0/s1 |
|---|
| CAS
number: |
97-67-6 |
|---|
| IUPAC Name: | (2S)-2-hydroxybutanedioate |
|---|
|
Traditional IUPAC Name: |
(-)-malic acid |
|---|
| SMILES: | C(=O)([O-])CC(O)C([O-])=O |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as beta hydroxy acids and derivatives. These are compounds containing a carboxylic acid substituted with a hydroxyl group on the C3 carbon atom. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
|
Class |
Hydroxy acids and derivatives |
|---|
| Sub Class | Beta hydroxy acids and derivatives |
|---|
|
Direct Parent |
Beta hydroxy acids and derivatives |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Short-chain hydroxy acid
- Beta-hydroxy acid
- Fatty acid
- Dicarboxylic acid or derivatives
- Secondary alcohol
- Carboxylic acid
- Carboxylic acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Carbonyl group
- Alcohol
- Organic anion
- Aliphatic acyclic compound
|
|---|
| Molecular Framework |
Aliphatic acyclic compounds |
|---|
| External Descriptors |
- C4-dicarboxylate (CHEBI:15595)
- \u003ci\u003eRS\u003c/i\u003e-malate (MAL)
|
|---|
|
Physical Properties |
|---|
| State: |
Solid |
|---|
| Charge: | -2 |
|---|
|
Melting point: |
107 °C |
|---|
| Experimental Properties: |
| Property | Value | Reference |
|---|
| Melting Point | 107 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Not Available |
|---|
| Reactions: | |
|---|
|
Pathways: |
|
|---|
|
Spectra |
|---|
| Spectra: |
| Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (3 TMS) | splash10-000t-0940000000-142d6f5fc2efbf0d5109 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (3 TMS) | splash10-0002-0920000000-505483ce10ee3c4c9d20 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (3 TMS) | splash10-00di-9710000000-8f263d045d715ae9fe2b | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS | Not Available |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-000i-9300000000-acd7d0159b8bb70b80fd | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-0096-9000000000-7359292556d9d393d8f4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0097-9000000000-a111335688b3219c1fc1 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-00lr-0942120000-9504700a82dcf5dc3adb | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-0a4i-0900000000-45e58d8a75d957cc5c41 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Negative | splash10-03di-0900000000-4ec56105367de7bf0027 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-001i-0900000000-1decf12f117200e0f28d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-03di-4900000000-86fd329658581c81c0ec | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-00di-9100000000-82ffef2aa053f77cdbc5 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-00di-9000000000-424bd54bc81a9db65a55 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-0006-9000000000-a916941735427ee3c48d | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | Not Available |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | Not Available |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | Not Available |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | Not Available |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | Not Available |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | Not Available |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available |
|---|
| 2D NMR | [1H,1H] 2D NMR Spectrum | Not Available |
|---|
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available |
|---|
|
|---|
|
References |
|---|
| References: |
- Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM: Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009 Feb 12;457(7231):910-4. [19212411 ]
- Shoemaker JD, Elliott WH: Automated screening of urine samples for carbohydrates, organic and amino acids after treatment with urease. J Chromatogr. 1991 Jan 2;562(1-2):125-38. [2026685 ]
- Zupke C, Sinskey AJ, Stephanopoulos G: Intracellular flux analysis applied to the effect of dissolved oxygen on hybridomas. Appl Microbiol Biotechnol. 1995 Dec;44(1-2):27-36. [8579834 ]
- Fang TJ, Dalmasso JP: Antimicrobial activity of sulfur dioxide to certain lactic acid bacteria isolated from wines. Zhonghua Min Guo Wei Sheng Wu Ji Mian Yi Xue Za Zhi. 1993 Aug;26(3):116-31. [7982367 ]
- Splittstoesser DF, McLellan MR, Churey JJ: Heat resistance of Escherichia coli O157:H7 in apple juice. J Food Prot. 1996 Mar;59(3):226-9. [10463437 ]
- Denayrolles M, Aigle M, Lonvaud-Funel A: Cloning and sequence analysis of the gene encoding Lactococcus lactis malolactic enzyme: relationships with malic enzymes. FEMS Microbiol Lett. 1994 Feb 1;116(1):79-86. [8132158 ]
|
|---|
| Synthesis Reference: |
McKenzie, Alex.; Plenderleith, H. J.; Walker, Nellie. Optical activation of racemic acid by d-malic acid. Journal of the Chemical Society, Transactions (1923), 123 2875-80. |
|---|
| Material Safety Data Sheet (MSDS) |
Download (PDF) |
|---|
|
Links |
|---|
| External Links: |
|
|---|