|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB110744 |
|---|
|
Identification |
|---|
| Name: |
4-hydroxyphenylpyruvate |
|---|
| Description: | A 2-oxo monocarboxylic acid anion obtained by removal of a proton from the carboxylic acid group of 3-(4-hydroxyphenyl)pyruvic acid. |
|---|
|
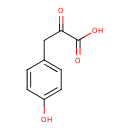
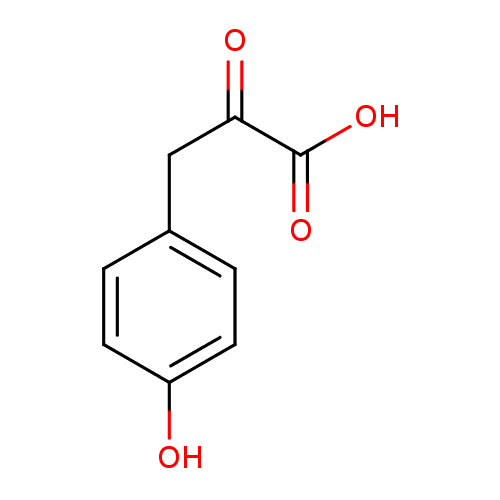
Structure |
|
|---|
| Synonyms: | -
p-hydroxyphenylpyruvic acid
-
3-(4-hydroxyphenyl)pyruvate
-
hydroxyphenylpyruvate
-
p-hydroxyphenylpyruvate
|
|---|
|
Chemical Formula: |
C9H7O4
|
|---|
| Average Molecular Weight: |
179.15 |
|---|
| Monoisotopic Molecular
Weight: |
180.0422587452 |
|---|
| InChI Key: |
KKADPXVIOXHVKN-UHFFFAOYSA-M |
|---|
| InChI: |
InChI=1S/C9H8O4/c10-7-3-1-6(2-4-7)5-8(11)9(12)13/h1-4,10H,5H2,(H,12,13)/p-1 |
|---|
| CAS
number: |
156-39-8 |
|---|
| IUPAC Name: | 3-(4-hydroxyphenyl)-2-oxopropanoate |
|---|
|
Traditional IUPAC Name: |
4-hydroxyphenylpyruvic acid |
|---|
| SMILES: | C1(C(CC(C([O-])=O)=O)=CC=C(C=1)O) |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of chemical entities known as phenylpyruvic acid derivatives. These are compounds containing a phenylpyruvic acid moiety, which consists of a phenyl group substituted at the second position by an pyruvic acid. |
|---|
|
Kingdom |
Chemical entities |
|---|
| Super Class | Organic compounds |
|---|
|
Class |
Benzenoids |
|---|
| Sub Class | Benzene and substituted derivatives |
|---|
|
Direct Parent |
Phenylpyruvic acid derivatives |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Phenylpyruvate
- 3-phenylpropanoic-acid
- 1-hydroxy-2-unsubstituted benzenoid
- Phenol
- Alpha-keto acid
- Keto acid
- Alpha-hydroxy ketone
- Ketone
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Organic oxide
- Organic oxygen compound
- Carbonyl group
- Hydrocarbon derivative
- Organooxygen compound
- Aromatic homomonocyclic compound
|
|---|
| Molecular Framework |
Aromatic homomonocyclic compounds |
|---|
| External Descriptors |
|
|---|
|
Physical Properties |
|---|
| State: |
Solid |
|---|
| Charge: | -1 |
|---|
|
Melting point: |
219 - 220 °C |
|---|
| Experimental Properties: |
| Property | Value | Reference |
|---|
| Melting Point | 219 - 220 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Not Available |
|---|
| Reactions: | |
|---|
|
Pathways: |
|
|---|
|
Spectra |
|---|
| Spectra: |
| Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) | splash10-002f-1920000000-75b07d9c09371340939f | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (3 TMS) | splash10-00xr-9340000000-087aad2497b3493d27c0 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (1 MEOX; 2 TMS) | splash10-002o-5910000000-fd1e55c84c79bfeca559 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (1 MEOX; 2 TMS) | splash10-002f-1941000000-2b4c87544657895fcaf6 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (1 MEOX; 3 TMS) | splash10-014i-3492100000-483bdcea11d60fe4d306 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (1 MEOX; 3 TMS) | splash10-014i-6791000000-ddc0a3b695cd1d0769f2 | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS | Not Available |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Negative (Annotated) | splash10-0a4i-0900000000-12033042c41b550bed42 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Negative (Annotated) | splash10-0a4i-0900000000-6e8a3701c6d254cc824d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Negative (Annotated) | splash10-0560-1900000000-8082c68e259d24234d05 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-004i-0900000000-2416d7f64101b9473cbb | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-0a4i-2900000000-b4c21b3d9751b9e56d67 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-0005-9400000000-708e258e692a0abfbc92 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-0002-9100000000-7d75e23ae6944c1e9c6d | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-001m-9000000000-fb80dcbab23323a042af | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Positive | splash10-08gr-0900000000-be3cb913118057f499d1 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Positive | splash10-06rj-0900000000-6bebea0fda2ef3a2263b | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Positive | splash10-0a6r-6900000000-ede11b81a97e891c66e3 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 10V, Negative | splash10-004i-0900000000-2584f872c980915c1739 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 20V, Negative | splash10-01s9-1900000000-249c71cfa9cefe2a8020 | View in MoNA |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 40V, Negative | splash10-001l-3900000000-66991d861a91be9b9286 | View in MoNA |
|---|
| MS | Mass Spectrum (Electron Ionization) | splash10-0a59-5900000000-6c003e26816579df31ca | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available |
|---|
| 2D NMR | [1H,1H] 2D NMR Spectrum | Not Available |
|---|
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available |
|---|
|
|---|
|
References |
|---|
| References: |
- Graham DE, Xu H, White RH (2003)Identification of the 7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase required for coenzyme F(420) biosynthesis. Archives of microbiology 180, Pubmed: 14593448
- Choi KP, Kendrick N, Daniels L (2002)Demonstration that fbiC is required by Mycobacterium bovis BCG for coenzyme F(420) and FO biosynthesis. Journal of bacteriology 184, Pubmed: 11948155
|
|---|
| Synthesis Reference: |
Billek, Gerhard. p-Hydroxyphenylpyruvic acid. Organic Syntheses (1963), 43 49-54. |
|---|
| Material Safety Data Sheet (MSDS) |
Download (PDF) |
|---|
|
Links |
|---|
| External Links: |
|
|---|