|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB110664 |
|---|
|
Identification |
|---|
| Name: |
L-lysine |
|---|
| Description: | An optically active form of lysinium having L-configuration. |
|---|
|
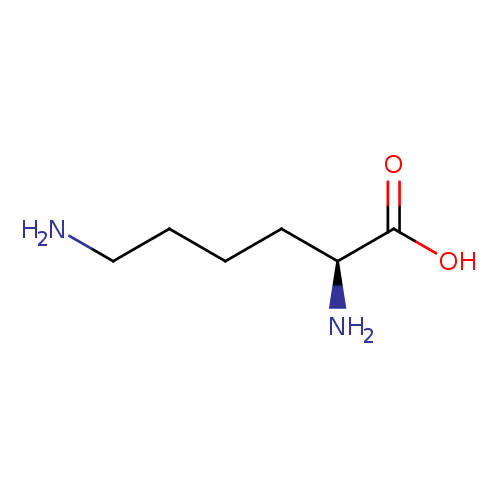
Structure |
|
|---|
| Synonyms: | -
K
-
lysine
-
L-lys
-
lys
-
2,6-diaminohexanoic acid
-
lysine acid
|
|---|
|
Chemical Formula: |
C6H15N2O2
|
|---|
| Average Molecular Weight: |
147.2 |
|---|
| Monoisotopic Molecular
Weight: |
148.1211777682 |
|---|
| InChI Key: |
KDXKERNSBIXSRK-YFKPBYRVSA-O |
|---|
| InChI: |
InChI=1S/C6H14N2O2/c7-4-2-1-3-5(8)6(9)10/h5H,1-4,7-8H2,(H,9,10)/p+1/t5-/m0/s1 |
|---|
| CAS
number: |
56-87-1 |
|---|
| IUPAC Name: | (2S)-2,6-diaminohexanoic acid |
|---|
|
Traditional IUPAC Name: |
L-lysine |
|---|
| SMILES: | C([N+])CCCC([N+])C([O-])=O |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Organic acids and derivatives |
|---|
|
Class |
Carboxylic acids and derivatives |
|---|
| Sub Class | Amino acids, peptides, and analogues |
|---|
|
Direct Parent |
L-alpha-amino acids |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- L-alpha-amino acid
- Medium-chain fatty acid
- Amino fatty acid
- Fatty acid
- Fatty acyl
- Amino acid
- Monocarboxylic acid or derivatives
- Carboxylic acid
- Organic oxide
- Organopnictogen compound
- Organic oxygen compound
- Primary amine
- Organooxygen compound
- Organonitrogen compound
- Primary aliphatic amine
- Carbonyl group
- Organic nitrogen compound
- Amine
- Hydrocarbon derivative
- Aliphatic acyclic compound
|
|---|
| Molecular Framework |
Aliphatic acyclic compounds |
|---|
| External Descriptors |
|
|---|
|
Physical Properties |
|---|
| State: |
Solid |
|---|
| Charge: | Not Available |
|---|
|
Melting point: |
224.5 °C |
|---|
| Experimental Properties: |
| Property | Value | Reference |
|---|
| Melting Point | 224.5 °C | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | 1000.0 mg/mL | Not Available | | LogP | -3.05 | HANSCH,C ET AL. (1995) |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Not Available |
|---|
| Reactions: | |
|---|
|
Pathways: |
|
|---|
|
Spectra |
|---|
| Spectra: |
| Spectrum Type | Description | Splash Key | |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (3 TMS) | splash10-00di-3910000000-98c565675de67aa87900 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) | splash10-0ab9-1910000000-87ef8534f592041f50f2 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-EI-TOF (Pegasus III TOF-MS system, Leco; GC 6890, Agilent Technologies) (4 TMS) | splash10-0a4i-1921000000-84f7815b0f650fa17444 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (3 TMS) | splash10-001i-9600000000-823408dba509cb204acf | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (3 TMS) | splash10-00di-3910000000-4f5578af5e7d8b6c49f7 | View in MoNA |
|---|
| GC-MS | GC-MS Spectrum - GC-MS (4 TMS) | splash10-0adi-1921000000-4e56d95e623e792f9e6b | View in MoNA |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - GC-MS | Not Available |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 10V, Positive (Annotated) | splash10-003r-8900000000-470a0beb4f338ed89bca | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 25V, Positive (Annotated) | splash10-001i-9000000000-74e9193d9d33c2509bfa | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - Quattro_QQQ 40V, Positive (Annotated) | splash10-0a59-9000000000-822c4e78250fffa56e39 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0002-0900000000-04f9a62a77fb5a37ca22 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-9000000000-035035ecfa084671479b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-5bb15839f86f4fca0d0b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-6c8ef03aa83eb1cab35b | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-01ot-0910000000-c182a7dcdbc260666978 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-0a4j-0900000000-6c5f378cef2f14204e15 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-001i-0900000000-41e1a6499097748934b0 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-ITFT (LTQ Orbitrap XL, Thermo Scientfic) , Positive | splash10-03di-0390000000-7270a0b85b9e3f9f6373 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Negative | splash10-0002-0900000000-a997e809874357908880 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Negative | splash10-0002-2900000000-f64110414f82c93e1fe8 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Negative | splash10-014m-9200000000-33a38e6370811c5ddc82 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Negative | splash10-0006-9000000000-3a8b0b6e62f5c66d3720 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Negative | splash10-0006-9000000000-d5167570d11d77fd541e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 10V, Positive | splash10-0002-0900000000-a46231bd529176101129 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 20V, Positive | splash10-001i-9200000000-f8b9f01b2a9886c51df9 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 30V, Positive | splash10-001i-9000000000-ace0361476d939043e98 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 40V, Positive | splash10-001i-9000000000-5b06b6e9dcf9faf8e89c | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QQ (API3000, Applied Biosystems) 50V, Positive | splash10-053r-9000000000-2894ef6d7f72ea71688e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - CE-ESI-TOF (CE-system connected to 6210 Time-of-Flight MS, Agilent) , Positive | splash10-0002-0900000000-290902f43cf851e8ef5e | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-001i-9300000000-f81193f6b50235ec8147 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Positive | splash10-001i-9300000000-8931d2193adab166d5e4 | View in MoNA |
|---|
| LC-MS/MS | LC-MS/MS Spectrum - LC-ESI-QTOF (UPLC Q-Tof Premier, Waters) , Negative | splash10-0002-0900000000-b4825cc64fcb830c6967 | View in MoNA |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available |
|---|
| 1D NMR | 13C NMR Spectrum | Not Available |
|---|
| 1D NMR | 1H NMR Spectrum | Not Available |
|---|
| 2D NMR | [1H,1H] 2D NMR Spectrum | Not Available |
|---|
| 2D NMR | [1H,13C] 2D NMR Spectrum | Not Available |
|---|
|
|---|
|
References |
|---|
| References: |
- Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, Nyati MK, Ahsan A, Kalyana-Sundaram S, Han B, Cao X, Byun J, Omenn GS, Ghosh D, Pennathur S, Alexander DC, Berger A, Shuster JR, Wei JT, Varambally S, Beecher C, Chinnaiyan AM: Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature. 2009 Feb 12;457(7231):910-4. [19212411 ]
- Silwood CJ, Lynch E, Claxson AW, Grootveld MC: 1H and (13)C NMR spectroscopic analysis of human saliva. J Dent Res. 2002 Jun;81(6):422-7. [12097436 ]
- Nicholson JK, O'Flynn MP, Sadler PJ, Macleod AF, Juul SM, Sonksen PH: Proton-nuclear-magnetic-resonance studies of serum, plasma and urine from fasting normal and diabetic subjects. Biochem J. 1984 Jan 15;217(2):365-75. [6696735 ]
- Engelborghs S, Marescau B, De Deyn PP: Amino acids and biogenic amines in cerebrospinal fluid of patients with Parkinson's disease. Neurochem Res. 2003 Aug;28(8):1145-50. [12834252 ]
- Hagenfeldt L, Bjerkenstedt L, Edman G, Sedvall G, Wiesel FA: Amino acids in plasma and CSF and monoamine metabolites in CSF: interrelationship in healthy subjects. J Neurochem. 1984 Mar;42(3):833-7. [6198473 ]
- Peng CT, Wu KH, Lan SJ, Tsai JJ, Tsai FJ, Tsai CH: Amino acid concentrations in cerebrospinal fluid in children with acute lymphoblastic leukemia undergoing chemotherapy. Eur J Cancer. 2005 May;41(8):1158-63. Epub 2005 Apr 14. [15911239 ]
- Cynober LA: Plasma amino acid levels with a note on membrane transport: characteristics, regulation, and metabolic significance. Nutrition. 2002 Sep;18(9):761-6. [12297216 ]
- Rainesalo S, Keranen T, Palmio J, Peltola J, Oja SS, Saransaari P: Plasma and cerebrospinal fluid amino acids in epileptic patients. Neurochem Res. 2004 Jan;29(1):319-24. [14992292 ]
- Kranz BR: Detection of rare malignant cells and their apoptotic fragments in cerebrospinal fluid. Lancet. 2000 Oct 7;356(9237):1242-4. [11072949 ]
- Hajishengallis G, Koga T, Russell MW: Affinity and specificity of the interactions between Streptococcus mutans antigen I/II and salivary components. J Dent Res. 1994 Sep;73(9):1493-502. [7523469 ]
- Pahler A, Parker J, Dekant W: Dose-dependent protein adduct formation in kidney, liver, and blood of rats and in human blood after perchloroethene inhalation. Toxicol Sci. 1999 Mar;48(1):5-13. [10330678 ]
- Faraasen S, Voros J, Csucs G, Textor M, Merkle HP, Walter E: Ligand-specific targeting of microspheres to phagocytes by surface modification with poly(L-lysine)-grafted poly(ethylene glycol) conjugate. Pharm Res. 2003 Feb;20(2):237-46. [12636162 ]
|
|---|
| Synthesis Reference: |
Rothstein, Morton. DL-Lysine-6-C14 and DL-a-aminoadipic acid-6-C14. Biochemical Preparations (1961), 8 85-8. |
|---|
| Material Safety Data Sheet (MSDS) |
Download (PDF) |
|---|
|
Links |
|---|
| External Links: |
|
|---|