|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB006672 |
|---|
|
Identification |
|---|
| Name: |
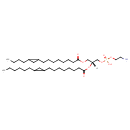
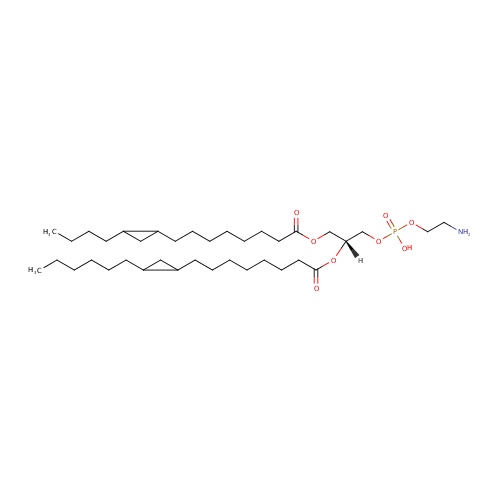
PE(15:0cyclo/17:0cycw7c) |
|---|
| Description: | PE(15:0cyclo/17:0cycw7c) is a phosphatidylethanolamine. It is a glycerophospholipid in which a phosphorylethanolamine moiety occupies a glycerol substitution site. As is the case with diacylglycerols, glycerophosphoethanolamines can have many different combinations of fatty acids of varying lengths and saturation attached to the C-1 and C-2 atoms. PE(15:0cyclo/17:0cycw7c), in particular, consists of one cis-9,10-Methylenetetradecanoic acid chain to the C-1 atom, and one heptadec-9-10-cyclo-anoyl to the C-2 atom. While most phospholipids have a saturated fatty acid on C-1 and an unsaturated fatty acid on C-2 of the glycerol backbone, the fatty acid distribution at the C-1 and C-2 positions of glycerol within phospholipids is continually in flux, owing to phospholipid degradation and the continuous phospholipid remodeling that occurs while these molecules are in membranes. PEs are neutral zwitterions at physiological pH. They mostly have palmitic or stearic acid on carbon 1 and a long chain unsaturated fatty acid (e.g. 18:2, 20:4 and 22:6) on carbon 2. PE synthesis can occur via two pathways. The first requires that ethanolamine be activated by phosphorylation and then coupled to CDP. The ethanolamine is then transferred from CDP-ethanolamine to phosphatidic acid to yield PE. The second involves the decarboxylation of PS. |
|---|
|
Structure |
|
|---|
| Synonyms: | Not Available |
|---|
|
Chemical Formula: |
C37H70NO8P |
|---|
| Average Molecular Weight: |
687.94 |
|---|
| Monoisotopic Molecular
Weight: |
687.483905216 |
|---|
| InChI Key: |
UUBQHHFVPKQSML-UASZDJHLSA-N |
|---|
| InChI: | InChI=1S/C37H70NO8P/c1-3-5-7-14-20-33-28-34(33)22-16-11-9-13-18-24-37(40)46-35(30-45-47(41,42)44-26-25-38)29-43-36(39)23-17-12-8-10-15-21-32-27-31(32)19-6-4-2/h31-35H,3-30,38H2,1-2H3,(H,41,42)/t31?,32?,33?,34?,35-/m1/s1 |
|---|
| CAS
number: |
Not Available |
|---|
| IUPAC Name: | (2-aminoethoxy)[(2R)-3-{[8-(2-butylcyclopropyl)octanoyl]oxy}-2-{[8-(2-hexylcyclopropyl)octanoyl]oxy}propoxy]phosphinic acid |
|---|
|
Traditional IUPAC Name: |
2-aminoethoxy((2R)-3-{[8-(2-butylcyclopropyl)octanoyl]oxy}-2-{[8-(2-hexylcyclopropyl)octanoyl]oxy}propoxy)phosphinic acid |
|---|
| SMILES: | CCCCCCC1CC1CCCCCCCC(=O)O[C@H](COC(=O)CCCCCCCC1CC1CCCC)COP(O)(=O)OCCN |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as phosphatidylethanolamines. These are glycerophosphoetahnolamines in which two fatty acids are bonded to the glycerol moiety through ester linkages. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
|
Class |
Glycerophospholipids |
|---|
| Sub Class | Glycerophosphoethanolamines |
|---|
|
Direct Parent |
Phosphatidylethanolamines |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Diacylglycero-3-phosphoethanolamine
- Phosphoethanolamine
- Dialkyl phosphate
- Fatty acid ester
- Fatty acyl
- Alkyl phosphate
- Phosphoric acid ester
- Dicarboxylic acid or derivatives
- Organic phosphoric acid derivative
- Organic phosphate
- Amino acid or derivatives
- Carboxylic acid ester
- Carboxylic acid derivative
- Amine
- Primary aliphatic amine
- Organonitrogen compound
- Organooxygen compound
- Primary amine
- Hydrocarbon derivative
- Carbonyl group
- Organic oxide
- Aliphatic homomonocyclic compound
|
|---|
| Molecular Framework |
Aliphatic homomonocyclic compounds |
|---|
| External Descriptors |
Not Available |
|---|
|
Physical Properties |
|---|
| State: |
Not Available |
|---|
| Charge: | 0 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Membrane |
|---|
| Reactions: | |
|---|
|
Pathways: |
Not Available |
|---|
|
Spectra |
|---|
| Spectra: |
|
|---|
|
References |
|---|
| References: |
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Yurtsever D. (2007). Fatty acid methyl ester profiling of Enterococcus and Esherichia coli for microbial source tracking. M.sc. Thesis. Villanova University: U.S.A
|
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
| Resource | Link |
|---|
| CHEBI ID | Not Available | | HMDB ID | Not Available | | Pubchem Compound ID | Not Available | | Kegg ID | Not Available | | ChemSpider ID | Not Available | | Wikipedia ID | Not Available | | BioCyc ID | Not Available |
|
|---|