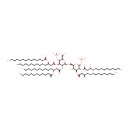|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB001507 |
|---|
|
Identification |
|---|
| Name: |
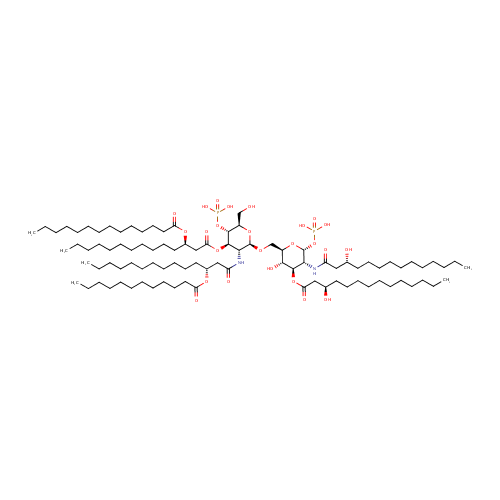
Lipid A |
|---|
| Description: | Lipid A is a lipid that comprises the innermost of the three regions of the lipopolysaccharide (LPS) molecule found in the outer membrane of Gram-negative bacteria such as Pseudomonas aeruginosa. Its hydrophobic nature allows it to anchor the LPS to the outer membrane. Lipid A consists of two glucosamine (carbohydrate/sugar) units with attached acyl chains ("fatty acids"), and normally containing one phosphate group on each carbohydrate. The Pseudomonas aeruginosa lipid A typically has four C14 hydroxy acyl chains attached to the sugars and one C14 and one C12 attached to the beta hydroxy groups. (Wikipedia) |
|---|
|
Structure |
|
|---|
| Synonyms: | - Diphosphoryl hexaacyl lipid A
- NULL
- Synthetic E. coli lipid A
|
|---|
|
Chemical Formula: |
C94H178N2O25P2 |
|---|
| Average Molecular Weight: |
1798.365 |
|---|
| Monoisotopic Molecular
Weight: |
1797.21939228 |
|---|
| InChI Key: |
GZQKNULLWNGMCW-PWQABINMSA-N |
|---|
| InChI: | InChI=1S/C94H178N2O25P2/c1-7-13-19-25-31-37-38-44-50-56-62-68-84(103)115-78(66-60-54-48-42-35-29-23-17-11-5)72-86(105)119-92-88(96-82(101)71-77(65-59-53-47-41-34-28-22-16-10-4)114-83(102)67-61-55-49-43-36-30-24-18-12-6)93(116-79(73-97)90(92)120-122(107,108)109)113-74-80-89(106)91(118-85(104)70-76(99)64-58-52-46-40-33-27-21-15-9-3)87(94(117-80)121-123(110,111)112)95-81(100)69-75(98)63-57-51-45-39-32-26-20-14-8-2/h75-80,87-94,97-99,106H,7-74H2,1-6H3,(H,95,100)(H,96,101)(H2,107,108,109)(H2,110,111,112)/t75-,76-,77-,78-,79-,80-,87-,88-,89-,90-,91-,92-,93-,94-/m1/s1 |
|---|
| CAS
number: |
Not Available |
|---|
| IUPAC Name: | {[(2R,3S,4R,5R,6R)-5-[(3R)-3-(dodecanoyloxy)tetradecanamido]-6-{[(2R,3S,4R,5R,6R)-3-hydroxy-5-[(3R)-3-hydroxytetradecanamido]-4-{[(3R)-3-hydroxytetradecanoyl]oxy}-6-(phosphonooxy)oxan-2-yl]methoxy}-2-(hydroxymethyl)-4-{[(3R)-3-(tetradecanoyloxy)tetradecanoyl]oxy}oxan-3-yl]oxy}phosphonic acid |
|---|
|
Traditional IUPAC Name: |
lipid A |
|---|
| SMILES: | CCCCCCCCCCCCCC(=O)O[C@H](CCCCCCCCCCC)CC(=O)O[C@@H]1[C@@H](NC(=O)C[C@@H](CCCCCCCCCCC)OC(=O)CCCCCCCCCCC)[C@H](OC[C@H]2O[C@H](OP(O)(O)=O)[C@H](NC(=O)C[C@H](O)CCCCCCCCCCC)[C@@H](OC(=O)C[C@H](O)CCCCCCCCCCC)[C@@H]2O)O[C@H](CO)[C@H]1OP(O)(O)=O |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as saccharolipids. These are compounds in which fatty acids are linked directly to a sugar backbone, forming structures that are compatible with membrane bilayers. In the saccharolipids, a sugar substitutes for the glycerol backbone that is present in glycerolipids and glycerophospholipids. The most familiar saccharolipids contain an acylated glucosamine. In contrast to others glycolipids, the fatty acid is not glycosidically linked to the sugar moiety. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
|
Class |
Saccharolipids |
|---|
| Sub Class | Not Available |
|---|
|
Direct Parent |
Saccharolipids |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Saccharolipid
- N-acyl-alpha-hexosamine
- Disaccharide phosphate
- Tetracarboxylic acid or derivatives
- Glucosamine
- Amino sugar
- O-glycosyl compound
- Glycosyl compound
- Disaccharide
- Monoalkyl phosphate
- Amino saccharide
- Fatty acid ester
- Beta-hydroxy acid
- Fatty acyl
- Alkyl phosphate
- Phosphoric acid ester
- Oxane
- Organic phosphoric acid derivative
- Organic phosphate
- Hydroxy acid
- Saccharide
- Secondary alcohol
- Carboxylic acid ester
- Oxacycle
- Organoheterocyclic compound
- Organic 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Carboxylic acid derivative
- Carboximidic acid derivative
- Carboximidic acid
- Acetal
- Hydrocarbon derivative
- Primary alcohol
- Organooxygen compound
- Organonitrogen compound
- Carbonyl group
- Alcohol
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework |
Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors |
|
|---|
|
Physical Properties |
|---|
| State: |
Solid |
|---|
| Charge: | -4 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Membrane |
|---|
| Reactions: | |
|---|
|
Pathways: |
- Lipopolysaccharide biosynthesis pae00540
|
|---|
|
Spectra |
|---|
| Spectra: |
|
|---|
|
References |
|---|
| References: |
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Uniprot Consortium (2012). "Reorganizing the protein space at the Universal Protein Resource (UniProt)." Nucleic Acids Res 40:D71-D75. Pubmed: 22102590
|
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|