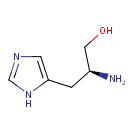
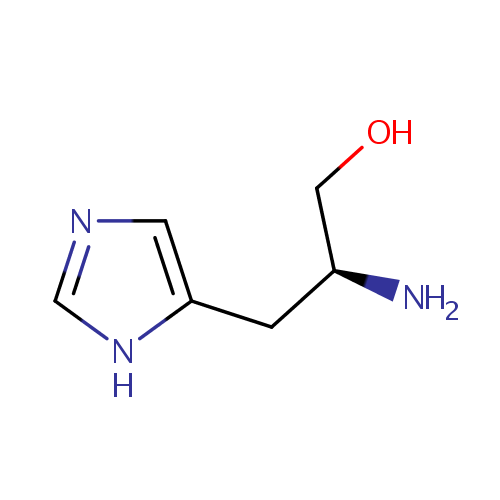
L-Histidinol (PAMDB000502)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000502 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | L-Histidinol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | L-Histidinol is a structural analogue of the essential amino acid L-histidine. It is an intermediate in histidine metabolism. It is converted to L-histidinal via bifunctional histidinal dehydrogenase/histidinol dehydrogenase (EC:1.1.1.23). (KEGG) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C6H11N3O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 141.171 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 141.090211989 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | ZQISRDCJNBUVMM-YFKPBYRVSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C6H11N3O/c7-5(3-10)1-6-2-8-4-9-6/h2,4-5,10H,1,3,7H2,(H,8,9)/t5-/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 4836-52-6 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S)-2-amino-3-(1H-imidazol-5-yl)propan-1-ol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | HSO | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | N[C@H](CO)CC1=CN=CN1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as aralkylamines. These are alkylamines in which the alkyl group is substituted at one carbon atom by an aromatic hydrocarbyl group. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organonitrogen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Amines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Aralkylamines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Aralkylamines | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteromonocyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Water + L-Histidinol + 2 NAD >3 Hydrogen ion + L-Histidine +2 NADH Water + Histidinol phosphate <> L-Histidinol + Phosphate L-Histidinol + NAD <> L-Histidinal + NADH + Hydrogen ion L-Histidinol + NAD > Hydrogen ion + histidinal + NADH Histidinol phosphate + Water > L-Histidinol + Inorganic phosphate L-Histidinol + Water + 2 NAD > L-Histidine +2 NADH L-histidinol-phosphate + Water > Phosphate + L-Histidinol | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Beltra, A. P.; Bonete, P.; Gonzalez-Garcia, J.; Garcia-Garcia, V.; Montiel, V. Electrochemical synthesis of L-histidinol using solvated electrons. Journal of the Electrochemical Society (2005), 152(4), D65-D68. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||