|
Record Information |
|---|
| Version |
1.0 |
|---|
| Update Date |
1/22/2018 11:54:54 AM |
|---|
|
Metabolite ID | PAMDB000403 |
|---|
|
Identification |
|---|
| Name: |
Queuine |
|---|
| Description: | Queuine is a member of the chemical class known as Aminocyclitols and Derivatives. These are cyclitols with at least one hydroxyl group replace by an amino group. Queuine (Q) is a hypermodified base found in the first (or wobble) position of the anticodon of tRNAs specific for Asn, Asp, His, and Tyr, in most eukaryotes and prokaryotes. The nucleoside of queuine is queuosine. Queuine is not found in the tRNA of archaea; however, a related 7-deazaguanine derivative, the nucleoside of which is archaeosine, occurs in different tRNA position, the dihydrouridine loop, and in tRNAs with more specificities. (WikiPedia) |
|---|
|
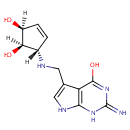
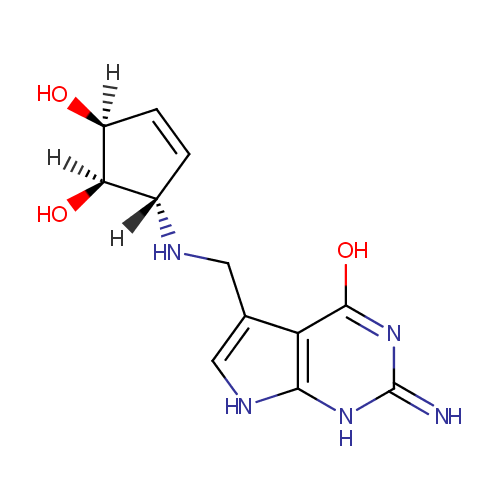
Structure |
|
|---|
| Synonyms: | - 2-Amino-5-((((1S,4S,5R)-4,5-dihydroxy-2-cyclopenten-1-yl)amino)methyl)-1,7-dihydro-4H-pyrrolo(2,3-D)pyrimidin-4-one
- 7-(3,4-trans-4,5-cis-Dihydroxy-1-cyclopenten-3-ylaminomethyl)-7-deazaguanine
- 7AMe7DAGua
- Base Q
- Queuine
|
|---|
|
Chemical Formula: |
C12H15N5O3 |
|---|
| Average Molecular Weight: |
277.2792 |
|---|
| Monoisotopic Molecular
Weight: |
277.117489371 |
|---|
| InChI Key: |
WYROLENTHWJFLR-BHNWBGBOSA-N |
|---|
| InChI: | InChI=1S/C12H15N5O3/c13-12-16-10-8(11(20)17-12)5(4-15-10)3-14-6-1-2-7(18)9(6)19/h1-2,4,6-7,9,14,18-19H,3H2,(H4,13,15,16,17,20)/t6-,7-,9+/m1/s1 |
|---|
| CAS
number: |
72496-59-4 |
|---|
| IUPAC Name: | (1R,2S,5S)-5-[({4-hydroxy-2-imino-1H,2H,7H-pyrrolo[2,3-d]pyrimidin-5-yl}methyl)amino]cyclopent-3-ene-1,2-diol |
|---|
|
Traditional IUPAC Name: |
(1R,2S,5S)-5-[({4-hydroxy-2-imino-1H,7H-pyrrolo[2,3-d]pyrimidin-5-yl}methyl)amino]cyclopent-3-ene-1,2-diol |
|---|
| SMILES: | O[C@@H]1C=C[C@@H](NCC2=CNC3=C2C(=O)NC(=N)N3)[C@@H]1O |
|---|
|
Chemical Taxonomy |
|---|
|
Taxonomy Description | This compound belongs to the class of organic compounds known as pyrrolopyrimidines. These are compounds containing a pyrrolopyrimidine moiety, which consists of a pyrrole ring fused to a pyrimidine. Pyrrole is 5-membered ring consisting of four carbon atoms and one nitrogen atom. Pyrimidine is a 6-membered ring consisting of four carbon atoms and two nitrogen centers at the 1- and 3- ring positions. |
|---|
|
Kingdom |
Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
|
Class |
Pyrrolopyrimidines |
|---|
| Sub Class | Not Available |
|---|
|
Direct Parent |
Pyrrolopyrimidines |
|---|
| Alternative Parents |
|
|---|
| Substituents |
- Pyrrolopyrimidine
- Hydroxypyrimidine
- Aralkylamine
- Substituted pyrrole
- Pyrimidine
- Heteroaromatic compound
- Pyrrole
- Cyclic alcohol
- Secondary alcohol
- 1,2-diol
- Azacycle
- Secondary amine
- Secondary aliphatic amine
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Amine
- Alcohol
- Aromatic heteropolycyclic compound
|
|---|
| Molecular Framework |
Aromatic heteropolycyclic compounds |
|---|
| External Descriptors |
|
|---|
|
Physical Properties |
|---|
| State: |
Not Available |
|---|
| Charge: | 2 |
|---|
|
Melting point: |
Not Available |
|---|
| Experimental Properties: |
|
|---|
| Predicted Properties |
|
|---|
|
Biological Properties |
|---|
| Cellular Locations: |
Cytoplasm |
|---|
| Reactions: | |
|---|
|
Pathways: |
Not Available |
|---|
|
Spectra |
|---|
| Spectra: |
|
|---|
|
References |
|---|
| References: |
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|
| Synthesis Reference: |
Not Available |
|---|
| Material Safety Data Sheet (MSDS) |
Not Available |
|---|
|
Links |
|---|
| External Links: |
|
|---|