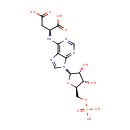
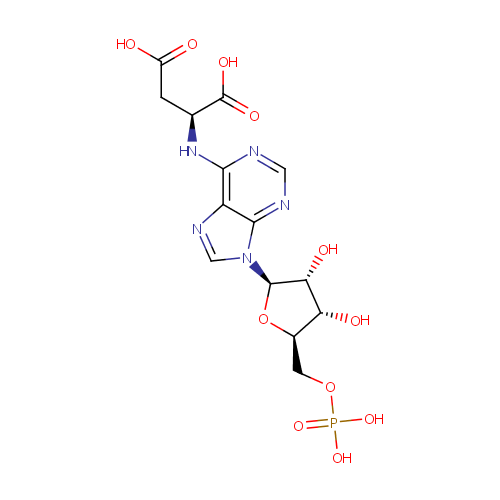
Adenylsuccinic acid (PAMDB000340)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000340 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Adenylsuccinic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Adenylsuccinic acid is a substrate of the enzyme adenylosuccinase (EC 4.3.2.2) in purine metabolism pathway. It is converted from IMP by adenylosuccinate synthetase (EC:6.3.4.4). In alanine, aspartate and glutamate metabolism pathway, it is converted from L-aspartate and a product of adenylosuccinate synthetase. (KEGG). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C14H18N5O11P | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 463.2934 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 463.074042955 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | OFBHPPMPBOJXRT-DPXQIYNJSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C14H18N5O11P/c20-7(21)1-5(14(24)25)18-11-8-12(16-3-15-11)19(4-17-8)13-10(23)9(22)6(30-13)2-29-31(26,27)28/h3-6,9-10,13,22-23H,1-2H2,(H,20,21)(H,24,25)(H,15,16,18)(H2,26,27,28)/t5?,6-,9-,10-,13-/m1/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 19046-78-7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S)-2-({9-[(2R,3R,4S,5R)-3,4-dihydroxy-5-[(phosphonooxy)methyl]oxolan-2-yl]-9H-purin-6-yl}amino)butanedioic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | adenylosuccinic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | O[C@@H]1[C@@H](COP(O)(O)=O)O[C@H]([C@@H]1O)N1C=NC2=C1N=CN=C2NC(CC(O)=O)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as purine ribonucleoside monophosphates. These are nucleotides consisting of a purine base linked to a ribose to which one monophosphate group is attached. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Nucleosides, nucleotides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Purine nucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Purine ribonucleotides | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Purine ribonucleoside monophosphates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aromatic heteropolycyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenylsuccinic acid <> Adenosine monophosphate + Fumaric acid L-Aspartic acid + Guanosine triphosphate + Inosinic acid <> Adenylsuccinic acid + Guanosine diphosphate +2 Hydrogen ion + Phosphate Guanosine triphosphate + Inosinic acid + L-Aspartic acid <> Guanosine diphosphate + Phosphate + Adenylsuccinic acid Adenylsuccinic acid + SAICAR <> Fumaric acid + Adenosine monophosphate + AICAR Inosinic acid + L-Aspartic acid + Guanosine triphosphate + L-Aspartic acid > Guanosine diphosphate + Phosphate +2 Hydrogen ion + N(6)-(1,2-dicarboxyethyl)AMP + Adenylsuccinic acid N(6)-(1,2-dicarboxyethyl)AMP + Adenylsuccinic acid > Fumaric acid + Adenosine monophosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Buck, Ildiko; Reese, Colin B. An unambiguous synthesis of adenylosuccinic acid and its constituent nucleoside. Journal of the Chemical Society, Perkin Transactions 1: Organic and Bio-Organic Chemistry (1972-1999) (1990), (11), 2937-42. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||