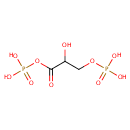
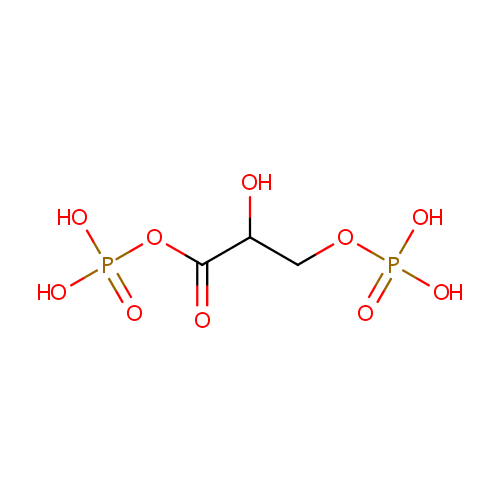
Glyceric acid 1,3-biphosphate (PAMDB000317)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000317 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Glyceric acid 1,3-biphosphate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | 1,3-Bisphosphogylcerate (1,3BPG), also known as PGAP, is a 3-carbon organic molecule present in most, if not all living creatures. It primarily exists as a metabolic intermediate in glycolysis during respiration. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C3H8O10P2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 266.0371 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 265.9592695 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | LJQLQCAXBUHEAZ-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C3H8O10P2/c4-2(1-12-14(6,7)8)3(5)13-15(9,10)11/h2,4H,1H2,(H2,6,7,8)(H2,9,10,11) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 1981-49-3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | {[2-hydroxy-3-(phosphonooxy)propanoyl]oxy}phosphonic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | 1,3-bisphosphoglycerate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OC(COP(O)(O)=O)C(=O)OP(O)(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as sugar acids and derivatives. These are compounds containing a saccharide unit which bears a carboxylic acid group. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organooxygen compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carbohydrates and carbohydrate conjugates | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Sugar acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Sugar acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | D-Glyceraldehyde 3-phosphate + NAD + Phosphate <> Glyceric acid 1,3-biphosphate + Hydrogen ion + NADH + 3-phospho-D-glyceroyl phosphate 3-Phosphoglycerate + Adenosine triphosphate <> Glyceric acid 1,3-biphosphate + ADP D-Glyceraldehyde 3-phosphate + Phosphate + NAD <> Glyceric acid 1,3-biphosphate + NADH + Hydrogen ion Adenosine triphosphate + 3-Phospho-D-glycerate <> ADP + Glyceric acid 1,3-biphosphate + 3-phospho-D-glyceroyl phosphate Glyceric acid 1,3-biphosphate + Water <> 3-Phospho-D-glycerate + Phosphate D-Glyceraldehyde 3-phosphate + NAD + Phosphate + D-Glyceraldehyde 3-phosphate > Glyceric acid 1,3-biphosphate + NADH + Hydrogen ion + Glyceric acid 1,3-biphosphate Glyceric acid 1,3-biphosphate + Adenosine diphosphate + Glyceric acid 1,3-biphosphate + ADP > Adenosine triphosphate + 3-Phosphoglyceric acid + 3-Phosphoglycerate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||