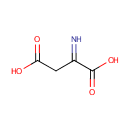
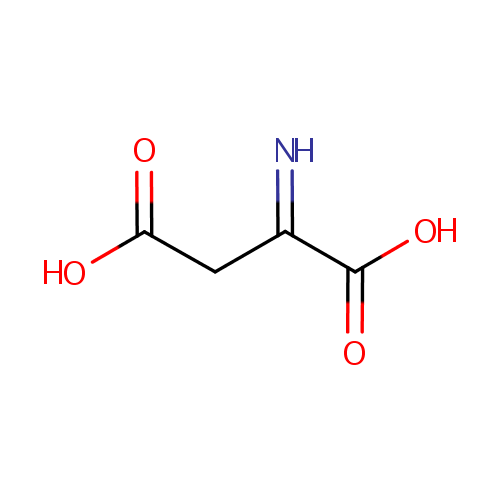
Iminoaspartic acid (PAMDB000267)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000267 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | Iminoaspartic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Iminoaspartic acid (alpha-iminosuccinate) is an intermediate in NAD biosynthesis from aspartate pathwyay. As a general rule, most prokaryotes utilize the aspartate de novo pathway, in which the nicotinate moiety of NAD is synthesized from aspartate, while in eukaryotes, the de novo pathway starts with tryptophan. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C4H5NO4 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 131.0868 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 131.021857653 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | NMUOATVLLQEYHI-UHFFFAOYSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C4H5NO4/c5-2(4(8)9)1-3(6)7/h5H,1H2,(H,6,7)(H,8,9) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 79067-61-1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | 2-iminobutanedioic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | iminosuccinic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | OC(=O)CC(=N)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as dicarboxylic acids and derivatives. These are organic compounds containing exactly two carboxylic acid groups. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Dicarboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | Dicarboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | -2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Dihydroxyacetone phosphate + Iminoaspartic acid <>2 Water + Phosphate + Quinolinic acid L-Aspartic acid + Fumaric acid > Hydrogen ion + Iminoaspartic acid + Succinic acid L-Aspartic acid + Oxygen <> Hydrogen ion + Hydrogen peroxide + Iminoaspartic acid L-Aspartic acid + Ubiquinone-8 > Hydrogen ion + Iminoaspartic acid + Ubiquinol-8 L-Aspartic acid + Menaquinone 8 > Hydrogen ion + Iminoaspartic acid + Menaquinol 8 L-Aspartic acid + Oxygen <> Iminoaspartic acid + Hydrogen peroxide Dihydroxyacetone phosphate + Iminoaspartic acid > Quinolinic acid +2 Water + Inorganic phosphate L-Aspartic acid + Oxygen + L-Aspartic acid > Hydrogen peroxide + Hydrogen ion + Iminoaspartic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Not Available | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||