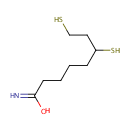
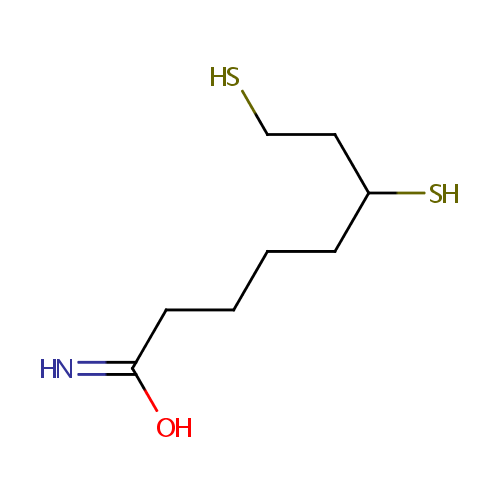
| References: |
- Brautigam CA, Chuang JL, Tomchick DR, Machius M, Chuang DT: Crystal structure of human dihydrolipoamide dehydrogenase: NAD+/NADH binding and the structural basis of disease-causing mutations. J Mol Biol. 2005 Jul 15;350(3):543-52. Pubmed: 15946682
- Deres P, Halmosi R, Toth A, Kovacs K, Palfi A, Habon T, Czopf L, Kalai T, Hideg K, Sumegi B, Toth K: Prevention of doxorubicin-induced acute cardiotoxicity by an experimental antioxidant compound. J Cardiovasc Pharmacol. 2005 Jan;45(1):36-43. Pubmed: 15613977
- Kanehisa, M., Goto, S., Sato, Y., Furumichi, M., Tanabe, M. (2012). "KEGG for integration and interpretation of large-scale molecular data sets." Nucleic Acids Res 40:D109-D114. Pubmed: 22080510
- Keseler, I. M., Collado-Vides, J., Santos-Zavaleta, A., Peralta-Gil, M., Gama-Castro, S., Muniz-Rascado, L., Bonavides-Martinez, C., Paley, S., Krummenacker, M., Altman, T., Kaipa, P., Spaulding, A., Pacheco, J., Latendresse, M., Fulcher, C., Sarker, M., Shearer, A. G., Mackie, A., Paulsen, I., Gunsalus, R. P., Karp, P. D. (2011). "EcoCyc: a comprehensive database of Escherichia coli biology." Nucleic Acids Res 39:D583-D590. Pubmed: 21097882
- Kim H: Asparagine-473 residue is important to the efficient function of human dihydrolipoamide dehydrogenase. J Biochem Mol Biol. 2005 Mar 31;38(2):248-52. Pubmed: 15826505
- Li XJ, Grunwald D, Mathieu J, Morel F, Stasia MJ: Crucial role of two potential cytosolic regions of Nox2, 191TSSTKTIRRS200 and 484DESQANHFAVHHDEEKD500, on NADPH oxidase activation. J Biol Chem. 2005 Apr 15;280(15):14962-73. Epub 2005 Jan 31. Pubmed: 15684431
- McMillan PJ, Stimmler LM, Foth BJ, McFadden GI, Muller S: The human malaria parasite Plasmodium falciparum possesses two distinct dihydrolipoamide dehydrogenases. Mol Microbiol. 2005 Jan;55(1):27-38. Pubmed: 15612914
- van der Werf, M. J., Overkamp, K. M., Muilwijk, B., Coulier, L., Hankemeier, T. (2007). "Microbial metabolomics: toward a platform with full metabolome coverage." Anal Biochem 370:17-25. Pubmed: 17765195
- Winder, C. L., Dunn, W. B., Schuler, S., Broadhurst, D., Jarvis, R., Stephens, G. M., Goodacre, R. (2008). "Global metabolic profiling of Escherichia coli cultures: an evaluation of methods for quenching and extraction of intracellular metabolites." Anal Chem 80:2939-2948. Pubmed: 18331064
|
|---|