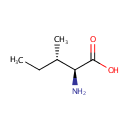
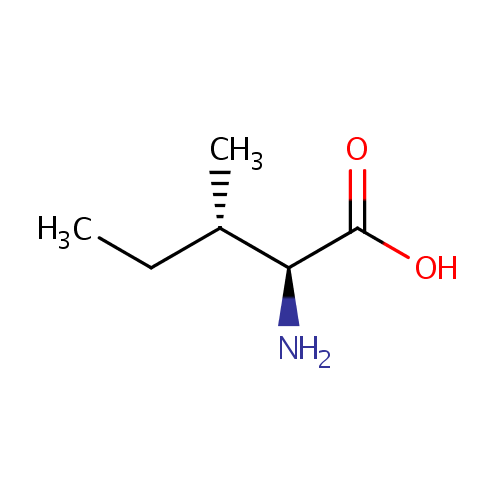
L-Isoleucine (PAMDB000071)
| Record Information | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Version | 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Update Date | 1/22/2018 11:54:54 AM | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Metabolite ID | PAMDB000071 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Identification | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Name: | L-Isoleucine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Description: | Isoleucine is an amino acid with the chemical formula HO2CCH(NH2)CH(CH3)CH2CH3. Its codons are AUU, AUC and AUA. With a hydrocarbon side chain, isoleucine is classified as a hydrophobic amino acid. Together with threonine, isoleucine is one of two common amino acids that have a chiral side chain. Four stereoisomers of isoleucine are possible, including two possible diastereomers of L-isoleucine. However, isoleucine present in nature exists in one enantiomeric form, (2S,3S)-2-amino-3-methylpentanoic acid. (Wikipedia) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Structure | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synonyms: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Formula: | C6H13NO2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Average Molecular Weight: | 131.1729 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Monoisotopic Molecular Weight: | 131.094628665 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI Key: | AGPKZVBTJJNPAG-WHFBIAKZSA-N | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| InChI: | InChI=1S/C6H13NO2/c1-3-4(2)5(7)6(8)9/h4-5H,3,7H2,1-2H3,(H,8,9)/t4-,5-/m0/s1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| CAS number: | 73-32-5 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| IUPAC Name: | (2S,3S)-2-amino-3-methylpentanoic acid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Traditional IUPAC Name: | L-isoleucine | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| SMILES: | CC[C@H](C)[C@H](N)C(O)=O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Chemical Taxonomy | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Taxonomy Description | This compound belongs to the class of organic compounds known as l-alpha-amino acids. These are alpha amino acids which have the L-configuration of the alpha-carbon atom. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Kingdom | Organic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Super Class | Organic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Class | Carboxylic acids and derivatives | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sub Class | Amino acids, peptides, and analogues | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Direct Parent | L-alpha-amino acids | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Alternative Parents | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Substituents |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Molecular Framework | Aliphatic acyclic compounds | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Descriptors |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Physical Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| State: | Solid | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Charge: | 0 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Melting point: | 285.5 °C | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Experimental Properties: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Predicted Properties |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Biological Properties | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Cellular Locations: | Cytoplasm | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reactions: | Adenosine triphosphate + Water + L-Isoleucine > ADP + Hydrogen ion + L-Isoleucine + Phosphate Adenosine triphosphate + Water + L-Isoleucine > ADP + Hydrogen ion + L-Isoleucine + Phosphate Adenosine triphosphate + L-Isoleucine + tRNA(Ile) + tRNA(Ile) <> Adenosine monophosphate + L-Isoleucyl-tRNA(Ile) + Pyrophosphate + L-Isoleucyl-tRNA(Ile) alpha-Ketoglutarate + L-Isoleucine <> 3-Methyl-2-oxovaleric acid + L-Glutamate Adenosine triphosphate + L-Isoleucine + tRNA(Ile) <> Adenosine monophosphate + Pyrophosphate + L-Isoleucyl-tRNA(Ile) -->-->L-Isoleucine + Oxoglutaric acid <> 3-Methyl-2-oxovaleric acid + L-Glutamate L-Isoleucine + Adenosine triphosphate + Hydrogen ion + tRNA(Ile) + L-Isoleucine > L-Isoleucyl-tRNA(Ile) + Adenosine monophosphate + Pyrophosphate 3-Methyl-2-oxovaleric acid + L-Glutamic acid + 3-Methyl-2-oxovaleric acid + L-Glutamate > Oxoglutaric acid + L-Isoleucine + L-Isoleucine L-Isoleucine + Adenosine triphosphate + Water + L-Isoleucine > L-Isoleucine + Adenosine diphosphate + Phosphate + Hydrogen ion + ADP | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Pathways: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Spectra: | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| References: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthesis Reference: | Marvel, C. S. L-Isoleucine. Organic Syntheses (1941), 21 60-4. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Material Safety Data Sheet (MSDS) | Download (PDF) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Links | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| External Links: |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Enzymes
- General function:
- Involved in nucleotide binding
- Specific function:
- Catalyzes the attachment of isoleucine to tRNA(Ile). As IleRS can inadvertently accommodate and process structurally similar amino acids such as valine, to avoid such errors it has two additional distinct tRNA(Ile)-dependent editing activities. One activity is designated as 'pretransfer' editing and involves the hydrolysis of activated Val-AMP. The other activity is designated 'posttransfer' editing and involves deacylation of mischarged Val-tRNA(Ile)
- Gene Name:
- ileS
- Locus Tag:
- PA4560
- Molecular weight:
- 105.5 kDa
Reactions
| ATP + L-isoleucine + tRNA(Ile) = AMP + diphosphate + L-isoleucyl-tRNA(Ile). |
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for branched-chain amino acids. Probably responsible for the translocation of the substrates across the membrane
- Gene Name:
- livH
- Locus Tag:
- PA1073
- Molecular weight:
- 32.5 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for branched-chain amino acids. Probably responsible for the translocation of the substrates across the membrane
- Gene Name:
- livM
- Locus Tag:
- PA1072
- Molecular weight:
- 45.6 kDa
- General function:
- Involved in catalytic activity
- Specific function:
- Acts on leucine, isoleucine and valine
- Gene Name:
- ilvE
- Locus Tag:
- PA5013
- Molecular weight:
- 34.1 kDa
Reactions
| L-leucine + 2-oxoglutarate = 4-methyl-2-oxopentanoate + L-glutamate. |
| L-isoleucine + 2-oxoglutarate = (S)-3-methyl-2-oxopentanoate + L-glutamate. |
| L-valine + 2-oxoglutarate = 3-methyl-2-oxobutanoate + L-glutamate. |
- General function:
- Involved in nucleotide binding
- Specific function:
- Component of the leucine-specific transport system
- Gene Name:
- livG
- Locus Tag:
- PA1071
- Molecular weight:
- 28.3 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Component of the leucine-specific transport system
- Gene Name:
- livF
- Locus Tag:
- PA1070
- Molecular weight:
- 25.6 kDa
Transporters
- General function:
- Involved in nucleotide binding
- Specific function:
- Probably part of a binding-protein-dependent transport system yecCS for an amino acid. Probably responsible for energy coupling to the transport system
- Gene Name:
- yecC
- Locus Tag:
- PA5152
- Molecular weight:
- 28.4 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for branched-chain amino acids. Probably responsible for the translocation of the substrates across the membrane
- Gene Name:
- livH
- Locus Tag:
- PA1073
- Molecular weight:
- 32.5 kDa
- General function:
- Involved in transporter activity
- Specific function:
- Part of the binding-protein-dependent transport system for branched-chain amino acids. Probably responsible for the translocation of the substrates across the membrane
- Gene Name:
- livM
- Locus Tag:
- PA1072
- Molecular weight:
- 45.6 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Component of the leucine-specific transport system
- Gene Name:
- livG
- Locus Tag:
- PA1071
- Molecular weight:
- 28.3 kDa
- General function:
- Involved in nucleotide binding
- Specific function:
- Component of the leucine-specific transport system
- Gene Name:
- livF
- Locus Tag:
- PA1070
- Molecular weight:
- 25.6 kDa
- General function:
- Involved in branched-chain aliphatic amino acid transmembrane transporter activity
- Specific function:
- Component of the LIV-II transport system for branched- chain amino acids. This LIV-II transport system may be H(+)- coupled
- Gene Name:
- brnQ
- Locus Tag:
- PA1971
- Molecular weight:
- 45.3 kDa